Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
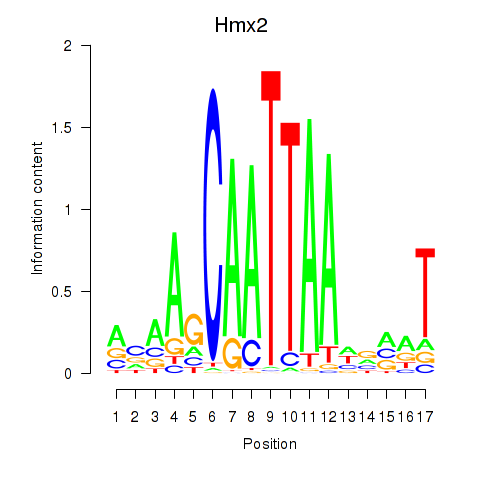
Results for Hmx2
Z-value: 0.74
Transcription factors associated with Hmx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx2
|
ENSMUSG00000050100.7 | H6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx2 | mm10_v2_chr7_+_131548755_131548773 | -0.02 | 9.3e-01 | Click! |
Activity profile of Hmx2 motif
Sorted Z-values of Hmx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_146079254 | 6.76 |
ENSMUST00000035571.8
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr19_-_7966000 | 5.60 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr13_-_23914998 | 4.58 |
ENSMUST00000021769.8
ENSMUST00000110407.2 |
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr2_-_24048857 | 3.03 |
ENSMUST00000114497.1
|
Hnmt
|
histamine N-methyltransferase |
| chr19_+_12633507 | 3.02 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr19_-_8405060 | 2.92 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr2_-_24049389 | 2.92 |
ENSMUST00000051416.5
|
Hnmt
|
histamine N-methyltransferase |
| chr19_+_12633303 | 2.76 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr13_+_4436094 | 2.65 |
ENSMUST00000156277.1
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr19_-_58454435 | 2.12 |
ENSMUST00000169850.1
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr19_-_8131982 | 1.97 |
ENSMUST00000065651.4
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr4_-_62054112 | 1.93 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr12_+_8771317 | 1.75 |
ENSMUST00000020911.7
|
Sdc1
|
syndecan 1 |
| chr5_-_146009598 | 1.70 |
ENSMUST00000138870.1
ENSMUST00000068317.6 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr7_+_119607014 | 1.51 |
ENSMUST00000126367.1
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr11_-_46389509 | 1.49 |
ENSMUST00000020664.6
|
Itk
|
IL2 inducible T cell kinase |
| chr16_-_57167307 | 1.41 |
ENSMUST00000023432.8
|
Nit2
|
nitrilase family, member 2 |
| chr3_+_94372794 | 1.41 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr5_+_8660059 | 1.39 |
ENSMUST00000047753.4
|
Abcb1a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1A |
| chr17_+_46428930 | 1.32 |
ENSMUST00000024764.5
ENSMUST00000165993.1 |
Crip3
|
cysteine-rich protein 3 |
| chr3_+_19957240 | 1.28 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr11_-_46389471 | 1.24 |
ENSMUST00000109237.2
|
Itk
|
IL2 inducible T cell kinase |
| chr17_+_46428917 | 1.18 |
ENSMUST00000113465.3
|
Crip3
|
cysteine-rich protein 3 |
| chr4_-_96553617 | 1.17 |
ENSMUST00000030303.5
|
Cyp2j6
|
cytochrome P450, family 2, subfamily j, polypeptide 6 |
| chr1_+_91298354 | 1.12 |
ENSMUST00000142488.1
ENSMUST00000124832.1 ENSMUST00000147523.1 |
Scly
|
selenocysteine lyase |
| chr9_-_15301555 | 1.10 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr1_-_162866502 | 1.09 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr19_+_29945782 | 1.08 |
ENSMUST00000025724.8
|
Il33
|
interleukin 33 |
| chr13_-_62888282 | 1.04 |
ENSMUST00000092888.4
|
Fbp1
|
fructose bisphosphatase 1 |
| chr2_+_167421706 | 1.02 |
ENSMUST00000047815.6
ENSMUST00000109218.1 ENSMUST00000073873.3 |
Slc9a8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8 |
| chrX_+_139563316 | 0.99 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr1_+_91298334 | 0.95 |
ENSMUST00000027532.6
|
Scly
|
selenocysteine lyase |
| chr2_-_75938407 | 0.90 |
ENSMUST00000099996.3
|
Ttc30b
|
tetratricopeptide repeat domain 30B |
| chr18_+_4993795 | 0.89 |
ENSMUST00000153016.1
|
Svil
|
supervillin |
| chr4_+_147492417 | 0.88 |
ENSMUST00000105721.2
|
Gm13152
|
predicted gene 13152 |
| chr12_+_8973892 | 0.86 |
ENSMUST00000085745.6
ENSMUST00000111113.2 |
Wdr35
|
WD repeat domain 35 |
| chr17_-_34628380 | 0.83 |
ENSMUST00000167097.2
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_-_19907645 | 0.82 |
ENSMUST00000166511.1
ENSMUST00000020182.8 |
Pex7
|
peroxisomal biogenesis factor 7 |
| chr4_-_128962420 | 0.82 |
ENSMUST00000119354.1
ENSMUST00000106068.1 ENSMUST00000030581.3 |
Adc
|
arginine decarboxylase |
| chr14_+_48446128 | 0.78 |
ENSMUST00000124720.1
|
Tmem260
|
transmembrane protein 260 |
| chr5_+_102768771 | 0.77 |
ENSMUST00000112852.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr11_-_106612928 | 0.75 |
ENSMUST00000042780.7
|
Tex2
|
testis expressed gene 2 |
| chr11_-_115037769 | 0.75 |
ENSMUST00000100239.2
|
Gm11711
|
predicted gene 11711 |
| chr3_+_19957037 | 0.74 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr1_-_172632931 | 0.72 |
ENSMUST00000027826.5
|
Dusp23
|
dual specificity phosphatase 23 |
| chr11_-_99374895 | 0.71 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr9_+_92457369 | 0.70 |
ENSMUST00000034941.7
|
Plscr4
|
phospholipid scramblase 4 |
| chr2_-_127444524 | 0.69 |
ENSMUST00000028848.3
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr10_-_19907545 | 0.67 |
ENSMUST00000134220.1
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr4_-_15149755 | 0.66 |
ENSMUST00000108273.1
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr14_+_48446340 | 0.63 |
ENSMUST00000111735.2
|
Tmem260
|
transmembrane protein 260 |
| chr7_-_45062393 | 0.60 |
ENSMUST00000129101.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr3_-_116712644 | 0.60 |
ENSMUST00000029569.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr9_-_122310921 | 0.58 |
ENSMUST00000180685.1
|
Gm26797
|
predicted gene, 26797 |
| chr11_-_99244058 | 0.58 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr6_-_113501818 | 0.58 |
ENSMUST00000101059.1
|
Prrt3
|
proline-rich transmembrane protein 3 |
| chr11_-_100822525 | 0.56 |
ENSMUST00000107358.2
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr10_+_121365078 | 0.55 |
ENSMUST00000040344.6
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr16_-_44332925 | 0.55 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr9_-_20728219 | 0.53 |
ENSMUST00000034692.7
|
Olfm2
|
olfactomedin 2 |
| chr14_-_51773578 | 0.53 |
ENSMUST00000073860.5
|
Ang4
|
angiogenin, ribonuclease A family, member 4 |
| chr9_-_50739365 | 0.52 |
ENSMUST00000117093.1
ENSMUST00000121634.1 |
Dixdc1
|
DIX domain containing 1 |
| chr8_+_71922810 | 0.52 |
ENSMUST00000119003.1
|
Zfp617
|
zinc finger protein 617 |
| chr5_-_131308076 | 0.52 |
ENSMUST00000160609.1
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 homolog (human) |
| chr16_-_44333135 | 0.52 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr8_-_22694061 | 0.52 |
ENSMUST00000131767.1
|
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr4_+_122836236 | 0.51 |
ENSMUST00000030412.4
ENSMUST00000121870.1 ENSMUST00000097902.4 |
Ppt1
|
palmitoyl-protein thioesterase 1 |
| chr8_+_25017211 | 0.51 |
ENSMUST00000033961.5
|
Tm2d2
|
TM2 domain containing 2 |
| chr5_+_29195983 | 0.51 |
ENSMUST00000160888.1
ENSMUST00000159272.1 ENSMUST00000001247.5 ENSMUST00000161398.1 ENSMUST00000160246.1 |
Rnf32
|
ring finger protein 32 |
| chr6_-_108185552 | 0.49 |
ENSMUST00000167338.1
ENSMUST00000172188.1 ENSMUST00000032191.9 |
Sumf1
|
sulfatase modifying factor 1 |
| chr18_+_62977854 | 0.47 |
ENSMUST00000150267.1
|
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr1_+_164275559 | 0.46 |
ENSMUST00000027867.6
|
Ccdc181
|
coiled-coil domain containing 181 |
| chr14_-_18893376 | 0.45 |
ENSMUST00000151926.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr2_-_26933781 | 0.44 |
ENSMUST00000154651.1
ENSMUST00000015011.3 |
Surf4
|
surfeit gene 4 |
| chr18_-_66022580 | 0.44 |
ENSMUST00000143990.1
|
Lman1
|
lectin, mannose-binding, 1 |
| chr3_+_106721672 | 0.43 |
ENSMUST00000098750.2
ENSMUST00000130105.1 |
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr18_-_34579072 | 0.42 |
ENSMUST00000079287.5
|
Nme5
|
NME/NM23 family member 5 |
| chr3_-_65392546 | 0.42 |
ENSMUST00000119896.1
|
Ssr3
|
signal sequence receptor, gamma |
| chr19_-_40402267 | 0.42 |
ENSMUST00000099467.3
ENSMUST00000099466.3 ENSMUST00000165212.1 ENSMUST00000165469.1 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr15_+_102407144 | 0.40 |
ENSMUST00000169619.1
|
Sp1
|
trans-acting transcription factor 1 |
| chr17_+_35841668 | 0.40 |
ENSMUST00000174124.1
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr19_+_58728887 | 0.40 |
ENSMUST00000048644.5
|
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr8_+_25808474 | 0.40 |
ENSMUST00000033979.4
|
Star
|
steroidogenic acute regulatory protein |
| chr14_+_73237891 | 0.39 |
ENSMUST00000044405.6
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr10_+_63061582 | 0.39 |
ENSMUST00000020266.8
ENSMUST00000178684.1 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr8_+_85432686 | 0.38 |
ENSMUST00000180883.1
|
1700051O22Rik
|
RIKEN cDNA 1700051O22 Gene |
| chr5_-_140830430 | 0.37 |
ENSMUST00000000153.4
|
Gna12
|
guanine nucleotide binding protein, alpha 12 |
| chr12_+_84451485 | 0.37 |
ENSMUST00000137170.1
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr1_+_164115264 | 0.37 |
ENSMUST00000162746.1
|
Selp
|
selectin, platelet |
| chr5_-_28467093 | 0.37 |
ENSMUST00000002708.3
|
Shh
|
sonic hedgehog |
| chr5_+_8893677 | 0.35 |
ENSMUST00000003717.8
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr8_+_90828820 | 0.35 |
ENSMUST00000109614.2
ENSMUST00000048665.6 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr19_-_57197377 | 0.34 |
ENSMUST00000111546.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chrX_-_164027965 | 0.34 |
ENSMUST00000033739.4
|
Car5b
|
carbonic anhydrase 5b, mitochondrial |
| chr11_+_99873389 | 0.34 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr4_+_33081505 | 0.34 |
ENSMUST00000147889.1
|
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr18_+_62977826 | 0.33 |
ENSMUST00000025474.6
|
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr1_-_57970104 | 0.33 |
ENSMUST00000114410.3
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr10_+_77613709 | 0.32 |
ENSMUST00000124024.2
|
Sumo3
|
SMT3 suppressor of mif two 3 homolog 3 (yeast) |
| chr5_+_145345254 | 0.32 |
ENSMUST00000079268.7
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr11_-_78080360 | 0.32 |
ENSMUST00000021183.3
|
Eral1
|
Era (G-protein)-like 1 (E. coli) |
| chr12_-_101913116 | 0.32 |
ENSMUST00000177536.1
ENSMUST00000176728.1 ENSMUST00000021605.7 |
Trip11
|
thyroid hormone receptor interactor 11 |
| chr11_+_58171648 | 0.32 |
ENSMUST00000020820.1
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chrX_-_56598069 | 0.32 |
ENSMUST00000059899.2
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr19_-_57197435 | 0.31 |
ENSMUST00000111550.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_-_56947411 | 0.31 |
ENSMUST00000107609.3
ENSMUST00000068792.6 |
Tmem245
|
transmembrane protein 245 |
| chr2_+_30392405 | 0.30 |
ENSMUST00000113612.3
ENSMUST00000123202.1 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr18_-_63387124 | 0.29 |
ENSMUST00000047480.6
ENSMUST00000046860.5 |
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr9_+_86485407 | 0.29 |
ENSMUST00000034987.8
|
Dopey1
|
dopey family member 1 |
| chr4_-_45532470 | 0.28 |
ENSMUST00000147448.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr12_-_40199315 | 0.28 |
ENSMUST00000095760.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr11_-_99351086 | 0.28 |
ENSMUST00000017732.2
|
Krt27
|
keratin 27 |
| chr6_-_129622685 | 0.28 |
ENSMUST00000032252.5
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr3_-_65392579 | 0.28 |
ENSMUST00000029414.5
|
Ssr3
|
signal sequence receptor, gamma |
| chr13_+_23684192 | 0.27 |
ENSMUST00000018246.4
|
Hist1h2bc
|
histone cluster 1, H2bc |
| chr2_-_17460610 | 0.26 |
ENSMUST00000145492.1
|
Nebl
|
nebulette |
| chr6_+_71282280 | 0.26 |
ENSMUST00000080949.7
|
Krcc1
|
lysine-rich coiled-coil 1 |
| chr12_-_98901478 | 0.26 |
ENSMUST00000065716.6
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr18_+_6765171 | 0.24 |
ENSMUST00000097680.5
|
Rab18
|
RAB18, member RAS oncogene family |
| chr12_-_90969768 | 0.24 |
ENSMUST00000181184.1
|
4930544I03Rik
|
RIKEN cDNA 4930544I03 gene |
| chr18_+_37819543 | 0.24 |
ENSMUST00000055935.5
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr4_+_145868794 | 0.24 |
ENSMUST00000123460.1
|
Gm13235
|
predicted gene 13235 |
| chr7_-_103788275 | 0.23 |
ENSMUST00000183254.1
|
Olfr67
|
olfactory receptor 67 |
| chr9_-_75611308 | 0.23 |
ENSMUST00000064433.3
|
Tmod2
|
tropomodulin 2 |
| chr19_-_57197496 | 0.22 |
ENSMUST00000111544.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr3_-_87748619 | 0.22 |
ENSMUST00000023846.4
|
Lrrc71
|
leucine rich repeat containing 71 |
| chr10_+_69925800 | 0.21 |
ENSMUST00000182029.1
|
Ank3
|
ankyrin 3, epithelial |
| chrM_+_7759 | 0.21 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr11_-_3931960 | 0.20 |
ENSMUST00000109990.1
ENSMUST00000020710.4 ENSMUST00000109989.3 ENSMUST00000109991.1 ENSMUST00000109993.2 |
Tcn2
|
transcobalamin 2 |
| chr2_-_45112890 | 0.20 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr16_+_17619503 | 0.20 |
ENSMUST00000165092.1
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr3_+_66985947 | 0.19 |
ENSMUST00000161726.1
ENSMUST00000160504.1 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr4_-_132398199 | 0.19 |
ENSMUST00000136711.1
ENSMUST00000084249.4 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr9_+_35423582 | 0.19 |
ENSMUST00000154652.1
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr9_-_54501496 | 0.19 |
ENSMUST00000118600.1
ENSMUST00000118163.1 |
Dmxl2
|
Dmx-like 2 |
| chrX_+_164373363 | 0.18 |
ENSMUST00000033751.7
|
Figf
|
c-fos induced growth factor |
| chr5_-_131307848 | 0.18 |
ENSMUST00000086023.5
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 homolog (human) |
| chr4_+_94739276 | 0.18 |
ENSMUST00000073939.6
ENSMUST00000102798.1 |
Tek
|
endothelial-specific receptor tyrosine kinase |
| chr7_-_78783026 | 0.17 |
ENSMUST00000032841.5
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr16_-_14291355 | 0.16 |
ENSMUST00000090287.3
|
Myh11
|
myosin, heavy polypeptide 11, smooth muscle |
| chr7_-_127449109 | 0.16 |
ENSMUST00000053392.4
|
Zfp689
|
zinc finger protein 689 |
| chr4_+_145670685 | 0.16 |
ENSMUST00000105738.2
|
Gm13242
|
predicted gene 13242 |
| chr10_+_69925766 | 0.15 |
ENSMUST00000182269.1
ENSMUST00000183261.1 ENSMUST00000183074.1 |
Ank3
|
ankyrin 3, epithelial |
| chr3_+_107090874 | 0.15 |
ENSMUST00000060946.3
ENSMUST00000182132.1 |
A930002I21Rik
|
RIKEN cDNA A930002I21 gene |
| chr4_+_146156824 | 0.15 |
ENSMUST00000168483.2
|
Zfp600
|
zinc finger protein 600 |
| chr7_-_127448993 | 0.14 |
ENSMUST00000106299.1
|
Zfp689
|
zinc finger protein 689 |
| chr5_-_28210168 | 0.14 |
ENSMUST00000117098.1
|
Cnpy1
|
canopy 1 homolog (zebrafish) |
| chr4_-_147702553 | 0.14 |
ENSMUST00000117638.1
|
Zfp534
|
zinc finger protein 534 |
| chr6_-_129385497 | 0.14 |
ENSMUST00000032261.6
|
Clec12b
|
C-type lectin domain family 12, member B |
| chr1_-_57970067 | 0.13 |
ENSMUST00000164963.1
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr5_+_114380591 | 0.13 |
ENSMUST00000074002.5
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr6_+_129045893 | 0.13 |
ENSMUST00000032257.7
|
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F |
| chr11_+_107479549 | 0.13 |
ENSMUST00000106750.4
|
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr5_-_28210022 | 0.12 |
ENSMUST00000118882.1
|
Cnpy1
|
canopy 1 homolog (zebrafish) |
| chrX_+_150594420 | 0.12 |
ENSMUST00000112713.2
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chrX_+_38316177 | 0.12 |
ENSMUST00000016471.2
ENSMUST00000115134.1 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr5_-_114380459 | 0.11 |
ENSMUST00000001125.5
|
Kctd10
|
potassium channel tetramerisation domain containing 10 |
| chr10_+_123264076 | 0.11 |
ENSMUST00000050756.7
|
Fam19a2
|
family with sequence similarity 19, member A2 |
| chr5_+_15516489 | 0.10 |
ENSMUST00000178227.1
|
Gm21847
|
predicted gene, 21847 |
| chr17_+_35841491 | 0.10 |
ENSMUST00000082337.6
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr3_+_121671317 | 0.10 |
ENSMUST00000098646.3
|
4930432M17Rik
|
RIKEN cDNA 4930432M17 gene |
| chr7_-_37772868 | 0.10 |
ENSMUST00000176205.1
|
Zfp536
|
zinc finger protein 536 |
| chr5_-_26089291 | 0.09 |
ENSMUST00000094946.4
|
Gm10471
|
predicted gene 10471 |
| chr4_+_145514884 | 0.09 |
ENSMUST00000105741.1
|
Gm13225
|
predicted gene 13225 |
| chr5_-_26004798 | 0.09 |
ENSMUST00000168875.1
|
Gm1979
|
predicted gene 1979 |
| chr5_+_100039990 | 0.09 |
ENSMUST00000169390.1
ENSMUST00000031268.6 |
Enoph1
|
enolase-phosphatase 1 |
| chr11_-_67771115 | 0.09 |
ENSMUST00000051765.8
|
Glp2r
|
glucagon-like peptide 2 receptor |
| chrX_-_134111708 | 0.09 |
ENSMUST00000159259.1
ENSMUST00000113275.3 |
Nox1
|
NADPH oxidase 1 |
| chr1_+_177729814 | 0.08 |
ENSMUST00000016106.5
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr18_-_65939048 | 0.08 |
ENSMUST00000025396.3
|
Rax
|
retina and anterior neural fold homeobox |
| chr1_+_46066738 | 0.08 |
ENSMUST00000069293.7
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr2_+_30982350 | 0.07 |
ENSMUST00000061544.4
ENSMUST00000138161.1 ENSMUST00000142232.1 |
Usp20
|
ubiquitin specific peptidase 20 |
| chr14_-_19823807 | 0.07 |
ENSMUST00000022341.5
|
2700060E02Rik
|
RIKEN cDNA 2700060E02 gene |
| chr4_-_26346882 | 0.07 |
ENSMUST00000041374.7
ENSMUST00000153813.1 |
Manea
|
mannosidase, endo-alpha |
| chr14_+_53428708 | 0.07 |
ENSMUST00000180549.1
|
Trav6-3
|
T cell receptor alpha variable 6-3 |
| chr4_-_88634411 | 0.07 |
ENSMUST00000134155.1
|
C87499
|
expressed sequence C87499 |
| chr14_-_52213379 | 0.07 |
ENSMUST00000140603.1
|
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr18_-_88894322 | 0.06 |
ENSMUST00000070116.5
ENSMUST00000125362.1 |
Socs6
|
suppressor of cytokine signaling 6 |
| chr8_-_110902442 | 0.06 |
ENSMUST00000041382.6
|
Fuk
|
fucokinase |
| chr9_-_113708209 | 0.06 |
ENSMUST00000111861.3
ENSMUST00000035086.6 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr5_-_25954344 | 0.05 |
ENSMUST00000162387.4
|
Gm21671
|
predicted gene, 21671 |
| chr2_-_151432392 | 0.05 |
ENSMUST00000127020.1
|
Gm14152
|
predicted gene 14152 |
| chr5_-_138272733 | 0.05 |
ENSMUST00000161665.1
ENSMUST00000100530.1 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr7_+_3645267 | 0.04 |
ENSMUST00000038913.9
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr14_+_52726612 | 0.04 |
ENSMUST00000184883.1
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr1_+_109993982 | 0.04 |
ENSMUST00000027542.6
|
Cdh7
|
cadherin 7, type 2 |
| chr13_-_113042243 | 0.04 |
ENSMUST00000099162.2
|
Gm10735
|
predicted gene 10735 |
| chr6_-_137169678 | 0.04 |
ENSMUST00000119610.1
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr1_+_53061637 | 0.03 |
ENSMUST00000027269.5
|
Mstn
|
myostatin |
| chr4_-_88634142 | 0.03 |
ENSMUST00000156062.1
ENSMUST00000107142.2 ENSMUST00000107143.1 ENSMUST00000053304.3 |
C87499
|
expressed sequence C87499 |
| chr1_-_173227229 | 0.03 |
ENSMUST00000049706.5
|
Fcer1a
|
Fc receptor, IgE, high affinity I, alpha polypeptide |
| chr4_+_146971976 | 0.03 |
ENSMUST00000146688.2
|
Gm13150
|
predicted gene 13150 |
| chr14_-_48667508 | 0.03 |
ENSMUST00000144465.1
ENSMUST00000133479.1 ENSMUST00000119070.1 ENSMUST00000152018.1 |
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chr3_+_108093109 | 0.03 |
ENSMUST00000151326.1
|
Gnat2
|
guanine nucleotide binding protein, alpha transducing 2 |
| chr2_-_33087169 | 0.03 |
ENSMUST00000102810.3
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr19_-_57197556 | 0.02 |
ENSMUST00000099294.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_+_99955715 | 0.02 |
ENSMUST00000102783.4
|
Pgm2
|
phosphoglucomutase 2 |
| chr3_-_64719602 | 0.02 |
ENSMUST00000168072.1
|
AC110575.1
|
vomeronasal 2, receptor 7 |
| chr14_+_52725299 | 0.02 |
ENSMUST00000181483.1
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr6_+_40471352 | 0.02 |
ENSMUST00000114779.2
ENSMUST00000031971.6 ENSMUST00000121360.1 ENSMUST00000117411.1 ENSMUST00000117830.1 |
Ssbp1
|
single-stranded DNA binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.9 | 2.7 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 10.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 1.8 | GO:0048627 | myoblast development(GO:0048627) |
| 0.5 | 2.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.3 | 1.4 | GO:0046618 | drug export(GO:0046618) |
| 0.3 | 1.9 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 1.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 1.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 1.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.2 | 1.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 1.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 2.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 1.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 0.4 | GO:1904339 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.6 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 1.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.3 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 0.6 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 2.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.4 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 4.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 6.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 1.0 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.5 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.4 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.8 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 1.4 | 5.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.9 | 2.7 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.7 | 10.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.6 | 1.9 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.5 | 2.1 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.4 | 4.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 1.7 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 1.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.3 | 1.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 2.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 1.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.3 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 1.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 5.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.6 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 7.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |