Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
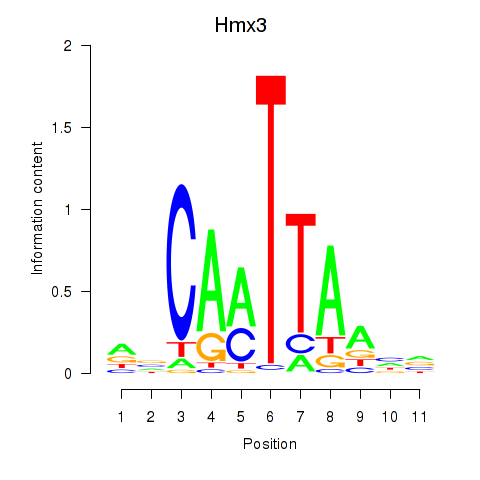
Results for Hmx3
Z-value: 0.29
Transcription factors associated with Hmx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx3
|
ENSMUSG00000040148.5 | H6 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx3 | mm10_v2_chr7_+_131542867_131542867 | -0.20 | 2.5e-01 | Click! |
Activity profile of Hmx3 motif
Sorted Z-values of Hmx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_20269162 | 0.87 |
ENSMUST00000024155.7
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr13_+_23746734 | 0.56 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chr10_-_80421847 | 0.50 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr15_+_66670749 | 0.45 |
ENSMUST00000065916.7
|
Tg
|
thyroglobulin |
| chr9_-_123678873 | 0.45 |
ENSMUST00000040960.6
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr9_-_123678782 | 0.44 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr7_+_24370255 | 0.39 |
ENSMUST00000171904.1
|
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr7_-_127208423 | 0.35 |
ENSMUST00000120705.1
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr4_+_154960915 | 0.27 |
ENSMUST00000049621.6
|
Hes5
|
hairy and enhancer of split 5 (Drosophila) |
| chr1_-_78968079 | 0.27 |
ENSMUST00000049117.5
|
Gm5830
|
predicted pseudogene 5830 |
| chr7_-_30559828 | 0.26 |
ENSMUST00000108164.1
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr6_+_86371489 | 0.25 |
ENSMUST00000089558.5
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr5_-_53707532 | 0.23 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr7_-_30559600 | 0.20 |
ENSMUST00000043975.4
ENSMUST00000156241.1 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr8_-_23237623 | 0.20 |
ENSMUST00000033950.5
|
Gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr4_-_129261394 | 0.18 |
ENSMUST00000145261.1
|
C77080
|
expressed sequence C77080 |
| chr11_+_69838514 | 0.16 |
ENSMUST00000133967.1
ENSMUST00000094065.4 |
Tmem256
|
transmembrane protein 256 |
| chr2_+_3114220 | 0.16 |
ENSMUST00000072955.5
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr11_+_114668524 | 0.16 |
ENSMUST00000106602.3
ENSMUST00000077915.3 ENSMUST00000106599.1 ENSMUST00000082092.4 |
Rpl38
|
ribosomal protein L38 |
| chr2_+_27165233 | 0.16 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chrX_+_150594420 | 0.16 |
ENSMUST00000112713.2
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr7_+_25681158 | 0.15 |
ENSMUST00000108403.3
|
B9d2
|
B9 protein domain 2 |
| chr19_+_41933464 | 0.15 |
ENSMUST00000026154.7
|
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr4_+_19818722 | 0.14 |
ENSMUST00000035890.7
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr3_-_155055341 | 0.13 |
ENSMUST00000064076.3
|
Tnni3k
|
TNNI3 interacting kinase |
| chr12_+_52699297 | 0.13 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_92375367 | 0.13 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr7_+_25267669 | 0.12 |
ENSMUST00000169266.1
|
Cic
|
capicua homolog (Drosophila) |
| chr9_+_19712829 | 0.12 |
ENSMUST00000077023.3
|
Olfr857
|
olfactory receptor 857 |
| chr2_-_153015331 | 0.11 |
ENSMUST00000028972.8
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr4_-_62470868 | 0.10 |
ENSMUST00000135811.1
ENSMUST00000120095.1 ENSMUST00000030087.7 ENSMUST00000107452.1 ENSMUST00000155522.1 |
Wdr31
|
WD repeat domain 31 |
| chr3_+_60006743 | 0.10 |
ENSMUST00000169794.1
|
Aadacl2
|
arylacetamide deacetylase-like 2 |
| chr3_+_96629919 | 0.10 |
ENSMUST00000048915.6
|
Rbm8a
|
RNA binding motif protein 8a |
| chr12_+_108179738 | 0.10 |
ENSMUST00000101055.4
|
Ccnk
|
cyclin K |
| chr14_-_55560340 | 0.09 |
ENSMUST00000066106.3
|
A730061H03Rik
|
RIKEN cDNA A730061H03 gene |
| chr4_+_80834123 | 0.09 |
ENSMUST00000133655.1
ENSMUST00000006151.6 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr18_-_37638719 | 0.09 |
ENSMUST00000058635.2
|
Slc25a2
|
solute carrier family 25 (mitochondrial carrier, ornithine transporter) member 2 |
| chr4_+_80834298 | 0.09 |
ENSMUST00000102831.1
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr11_+_103133333 | 0.08 |
ENSMUST00000124928.1
ENSMUST00000062530.4 |
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr2_+_57997884 | 0.08 |
ENSMUST00000112616.1
ENSMUST00000166729.1 |
Galnt5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 |
| chr4_+_144893127 | 0.08 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr1_-_158814469 | 0.08 |
ENSMUST00000161589.2
|
Pappa2
|
pappalysin 2 |
| chr7_-_45510400 | 0.08 |
ENSMUST00000033096.7
|
Nucb1
|
nucleobindin 1 |
| chr12_-_104473236 | 0.07 |
ENSMUST00000021513.4
|
Gsc
|
goosecoid homeobox |
| chr11_+_103133303 | 0.07 |
ENSMUST00000107037.1
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chrM_+_9452 | 0.07 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr2_+_130424321 | 0.07 |
ENSMUST00000128994.1
ENSMUST00000028900.9 |
Vps16
|
vacuolar protein sorting 16 (yeast) |
| chr5_-_137684665 | 0.07 |
ENSMUST00000100544.4
ENSMUST00000031736.9 ENSMUST00000151839.1 |
Agfg2
|
ArfGAP with FG repeats 2 |
| chr15_+_9436028 | 0.07 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chrX_+_159708593 | 0.06 |
ENSMUST00000080394.6
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr1_-_175979114 | 0.06 |
ENSMUST00000104983.1
|
B020018G12Rik
|
RIKEN cDNA B020018G12 gene |
| chr19_+_8802486 | 0.06 |
ENSMUST00000172175.1
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr1_+_110099295 | 0.06 |
ENSMUST00000134301.1
|
Cdh7
|
cadherin 7, type 2 |
| chr7_-_126897424 | 0.06 |
ENSMUST00000120007.1
|
Tmem219
|
transmembrane protein 219 |
| chr11_-_99374895 | 0.06 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr17_-_57247632 | 0.05 |
ENSMUST00000005975.6
|
Gpr108
|
G protein-coupled receptor 108 |
| chr7_+_3645267 | 0.05 |
ENSMUST00000038913.9
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr14_+_54464141 | 0.04 |
ENSMUST00000022782.8
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr14_+_52769753 | 0.04 |
ENSMUST00000178768.2
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr4_+_65604984 | 0.04 |
ENSMUST00000050850.7
ENSMUST00000107366.1 |
Trim32
|
tripartite motif-containing 32 |
| chr17_-_35697971 | 0.04 |
ENSMUST00000146472.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr11_-_99337930 | 0.04 |
ENSMUST00000100482.2
|
Krt26
|
keratin 26 |
| chr3_-_142169311 | 0.03 |
ENSMUST00000106230.1
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr2_-_84715160 | 0.03 |
ENSMUST00000035840.5
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr5_+_121777929 | 0.03 |
ENSMUST00000160821.1
|
Atxn2
|
ataxin 2 |
| chr15_+_101473472 | 0.03 |
ENSMUST00000088049.3
|
Krt86
|
keratin 86 |
| chr7_-_121035096 | 0.03 |
ENSMUST00000065740.2
|
Gm9905
|
predicted gene 9905 |
| chr2_-_72986716 | 0.03 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chr17_-_35046539 | 0.03 |
ENSMUST00000007250.7
|
Msh5
|
mutS homolog 5 (E. coli) |
| chr12_-_83487708 | 0.02 |
ENSMUST00000177959.1
ENSMUST00000178756.1 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr4_-_45532470 | 0.02 |
ENSMUST00000147448.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.2 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.2 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.2 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |