Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
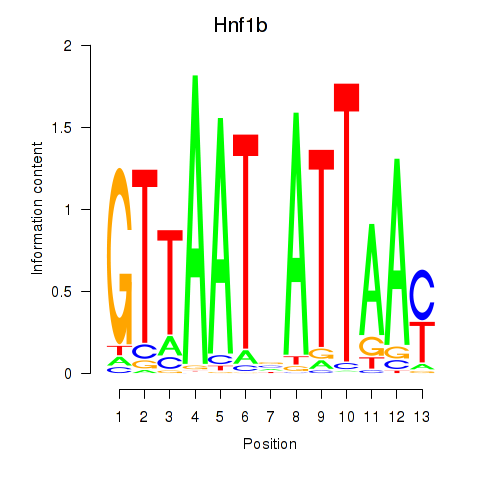
Results for Hnf1b
Z-value: 2.40
Transcription factors associated with Hnf1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf1b
|
ENSMUSG00000020679.5 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf1b | mm10_v2_chr11_+_83850832_83850855 | 0.07 | 6.9e-01 | Click! |
Activity profile of Hnf1b motif
Sorted Z-values of Hnf1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87337165 | 25.70 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr11_-_84167466 | 25.05 |
ENSMUST00000050771.7
|
Gm11437
|
predicted gene 11437 |
| chr8_-_5105232 | 24.67 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr5_-_89457763 | 23.29 |
ENSMUST00000049209.8
|
Gc
|
group specific component |
| chr9_-_48605147 | 20.82 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr15_+_10215955 | 19.12 |
ENSMUST00000130720.1
|
Prlr
|
prolactin receptor |
| chr13_-_23914998 | 18.89 |
ENSMUST00000021769.8
ENSMUST00000110407.2 |
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr1_-_130661584 | 17.91 |
ENSMUST00000137276.2
|
C4bp
|
complement component 4 binding protein |
| chr1_-_130661613 | 17.54 |
ENSMUST00000027657.7
|
C4bp
|
complement component 4 binding protein |
| chr11_-_78422217 | 16.97 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr11_+_101367542 | 16.79 |
ENSMUST00000019469.2
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr13_+_4233730 | 15.96 |
ENSMUST00000081326.6
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr2_-_134554348 | 14.49 |
ENSMUST00000028704.2
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr1_+_88211956 | 14.47 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr4_+_138967112 | 14.23 |
ENSMUST00000116094.2
|
Rnf186
|
ring finger protein 186 |
| chr19_+_30232921 | 13.57 |
ENSMUST00000025797.5
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr4_-_104876383 | 13.55 |
ENSMUST00000064873.8
ENSMUST00000106808.3 ENSMUST00000048947.8 |
C8a
|
complement component 8, alpha polypeptide |
| chr1_+_181051232 | 13.12 |
ENSMUST00000036819.6
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr16_+_22951072 | 13.05 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr1_+_88070765 | 12.67 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr1_+_88166004 | 12.59 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr19_-_46672883 | 12.51 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr13_+_23807027 | 12.27 |
ENSMUST00000006786.4
ENSMUST00000099697.2 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr5_-_87254804 | 12.02 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr13_-_4279420 | 11.95 |
ENSMUST00000021632.3
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr5_+_90518932 | 11.42 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr5_-_87092546 | 11.03 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr2_-_35100677 | 10.86 |
ENSMUST00000045776.4
ENSMUST00000113050.3 |
AI182371
|
expressed sequence AI182371 |
| chr13_+_4191163 | 10.81 |
ENSMUST00000021634.2
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr1_+_88138364 | 10.45 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr2_-_62412219 | 10.28 |
ENSMUST00000047812.7
|
Dpp4
|
dipeptidylpeptidase 4 |
| chr4_-_108144986 | 9.99 |
ENSMUST00000130776.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr3_+_129836729 | 9.95 |
ENSMUST00000077918.5
|
Cfi
|
complement component factor i |
| chr5_-_87569023 | 9.77 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr5_-_87535113 | 9.53 |
ENSMUST00000120150.1
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr17_+_12378603 | 9.14 |
ENSMUST00000014578.5
|
Plg
|
plasminogen |
| chr11_-_11898044 | 8.91 |
ENSMUST00000066237.3
|
Ddc
|
dopa decarboxylase |
| chr11_-_11898092 | 8.78 |
ENSMUST00000178704.1
|
Ddc
|
dopa decarboxylase |
| chr2_-_86347764 | 8.72 |
ENSMUST00000099894.2
|
Olfr1055
|
olfactory receptor 1055 |
| chr13_+_93674403 | 8.50 |
ENSMUST00000048001.6
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr1_+_88055377 | 8.39 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr13_+_23839445 | 8.24 |
ENSMUST00000091698.4
ENSMUST00000110422.1 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr1_+_88055467 | 8.24 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr16_+_43247278 | 8.17 |
ENSMUST00000114691.1
ENSMUST00000079441.6 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr13_+_23839401 | 8.10 |
ENSMUST00000039721.7
ENSMUST00000166467.1 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr5_+_90460889 | 7.98 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr4_-_140617062 | 7.94 |
ENSMUST00000154979.1
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr6_+_90333270 | 7.92 |
ENSMUST00000164761.1
ENSMUST00000046128.9 |
Uroc1
|
urocanase domain containing 1 |
| chr12_+_59095653 | 7.81 |
ENSMUST00000021384.4
|
Mia2
|
melanoma inhibitory activity 2 |
| chr7_+_119561699 | 7.62 |
ENSMUST00000167935.2
ENSMUST00000130583.1 |
Acsm2
|
acyl-CoA synthetase medium-chain family member 2 |
| chr15_-_50890396 | 7.57 |
ENSMUST00000185183.1
|
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr2_-_35061431 | 7.39 |
ENSMUST00000028233.3
|
Hc
|
hemolytic complement |
| chr3_+_115080965 | 6.92 |
ENSMUST00000051309.8
|
Olfm3
|
olfactomedin 3 |
| chr10_-_89533550 | 6.85 |
ENSMUST00000105297.1
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr11_+_70519619 | 6.81 |
ENSMUST00000179000.1
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr11_-_73326807 | 6.77 |
ENSMUST00000134079.1
|
Aspa
|
aspartoacylase |
| chr3_-_138131356 | 6.68 |
ENSMUST00000029805.8
|
Mttp
|
microsomal triglyceride transfer protein |
| chr3_-_98564038 | 6.63 |
ENSMUST00000058728.5
|
Gm10681
|
predicted gene 10681 |
| chr2_+_163547148 | 6.58 |
ENSMUST00000109411.1
ENSMUST00000018094.6 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr9_+_21835506 | 6.26 |
ENSMUST00000058777.6
|
Gm6484
|
predicted gene 6484 |
| chr7_-_119895446 | 6.25 |
ENSMUST00000098080.2
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr1_-_20617992 | 6.20 |
ENSMUST00000088448.5
|
Pkhd1
|
polycystic kidney and hepatic disease 1 |
| chr10_+_4611971 | 6.00 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr13_-_93674300 | 5.97 |
ENSMUST00000015941.7
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr2_+_162987330 | 5.95 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr4_-_6275629 | 5.88 |
ENSMUST00000029905.1
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr2_+_162987502 | 5.85 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr11_-_73326472 | 5.80 |
ENSMUST00000155630.2
|
Aspa
|
aspartoacylase |
| chr4_+_150853919 | 5.78 |
ENSMUST00000073600.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr16_+_96361654 | 5.77 |
ENSMUST00000113794.1
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr11_+_78465697 | 5.71 |
ENSMUST00000001126.3
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr9_-_71163224 | 5.60 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chr10_-_89621253 | 5.53 |
ENSMUST00000020102.7
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr19_+_43782181 | 5.53 |
ENSMUST00000026208.4
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr5_+_102845007 | 5.41 |
ENSMUST00000070000.4
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr13_+_89540636 | 5.21 |
ENSMUST00000022108.7
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr3_-_116712696 | 5.20 |
ENSMUST00000169530.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr3_+_14480692 | 5.14 |
ENSMUST00000037321.7
ENSMUST00000120484.1 ENSMUST00000120801.1 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr16_+_96361749 | 5.09 |
ENSMUST00000000163.6
ENSMUST00000081093.3 ENSMUST00000113795.1 |
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr13_+_55152640 | 5.02 |
ENSMUST00000005452.5
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr5_-_108675569 | 4.97 |
ENSMUST00000051757.7
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr15_+_99393574 | 4.92 |
ENSMUST00000162624.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr12_+_76533540 | 4.91 |
ENSMUST00000075249.4
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chrX_+_101376359 | 4.86 |
ENSMUST00000119080.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr1_+_133246092 | 4.85 |
ENSMUST00000038295.8
ENSMUST00000105082.2 |
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr7_-_45103747 | 4.85 |
ENSMUST00000003512.7
|
Fcgrt
|
Fc receptor, IgG, alpha chain transporter |
| chr10_+_61089327 | 4.73 |
ENSMUST00000020298.6
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| chr15_+_99393610 | 4.73 |
ENSMUST00000159531.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_+_99030946 | 4.57 |
ENSMUST00000030280.6
|
Angptl3
|
angiopoietin-like 3 |
| chr7_-_27166732 | 4.44 |
ENSMUST00000080058.4
|
Egln2
|
EGL nine homolog 2 (C. elegans) |
| chr4_+_48279794 | 4.36 |
ENSMUST00000030029.3
|
Invs
|
inversin |
| chr11_+_108395288 | 4.31 |
ENSMUST00000000049.5
|
Apoh
|
apolipoprotein H |
| chr2_-_164221605 | 4.22 |
ENSMUST00000018355.4
ENSMUST00000109376.2 |
Wfdc15b
|
WAP four-disulfide core domain 15B |
| chr5_+_31297551 | 4.21 |
ENSMUST00000072228.5
|
Gckr
|
glucokinase regulatory protein |
| chr15_-_55113460 | 4.17 |
ENSMUST00000100659.2
ENSMUST00000110230.1 |
Gm9920
|
predicted gene 9920 |
| chr11_+_70519183 | 4.13 |
ENSMUST00000057685.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr19_+_42045792 | 3.89 |
ENSMUST00000172244.1
ENSMUST00000081714.4 |
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr7_-_127895578 | 3.88 |
ENSMUST00000033074.6
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr11_+_4883186 | 3.85 |
ENSMUST00000139737.1
|
Nipsnap1
|
4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (C. elegans) |
| chr16_+_24448082 | 3.80 |
ENSMUST00000078988.2
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_78037931 | 3.79 |
ENSMUST00000072289.5
ENSMUST00000100784.2 |
Flot2
|
flotillin 2 |
| chr5_+_35041539 | 3.76 |
ENSMUST00000030985.6
|
Hgfac
|
hepatocyte growth factor activator |
| chr2_+_163506808 | 3.71 |
ENSMUST00000143911.1
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chrX_+_140907602 | 3.69 |
ENSMUST00000033806.4
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr12_+_4769375 | 3.68 |
ENSMUST00000178879.1
|
Pfn4
|
profilin family, member 4 |
| chr17_+_74395607 | 3.60 |
ENSMUST00000179074.1
ENSMUST00000024870.6 |
Slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr9_+_72958785 | 3.53 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr5_-_38491948 | 3.49 |
ENSMUST00000129099.1
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr5_-_114444036 | 3.48 |
ENSMUST00000031560.7
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) type B homolog (human) |
| chr19_+_5878622 | 3.46 |
ENSMUST00000136833.1
ENSMUST00000141362.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr9_+_54538984 | 3.45 |
ENSMUST00000060242.5
ENSMUST00000118413.1 |
Sh2d7
|
SH2 domain containing 7 |
| chr1_-_136960427 | 3.41 |
ENSMUST00000027649.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr19_+_60889749 | 3.40 |
ENSMUST00000003313.8
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr5_-_114443993 | 3.38 |
ENSMUST00000112245.1
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) type B homolog (human) |
| chr13_+_66932802 | 3.37 |
ENSMUST00000021990.3
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr10_+_63061582 | 3.36 |
ENSMUST00000020266.8
ENSMUST00000178684.1 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr9_+_120149733 | 3.33 |
ENSMUST00000068698.7
ENSMUST00000093773.1 ENSMUST00000111627.1 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr1_+_133309778 | 3.27 |
ENSMUST00000094557.4
ENSMUST00000183457.1 ENSMUST00000183738.1 ENSMUST00000185157.1 ENSMUST00000184603.1 |
Golt1a
Kiss1
GOLT1A
|
golgi transport 1 homolog A (S. cerevisiae) KiSS-1 metastasis-suppressor KISS1 isoform e |
| chr6_+_73248382 | 3.26 |
ENSMUST00000064740.6
|
Suclg1
|
succinate-CoA ligase, GDP-forming, alpha subunit |
| chr3_+_154597352 | 3.19 |
ENSMUST00000140644.1
ENSMUST00000144764.1 ENSMUST00000155232.1 |
Cryz
|
crystallin, zeta |
| chr3_+_90537242 | 3.17 |
ENSMUST00000098911.3
|
S100a16
|
S100 calcium binding protein A16 |
| chr3_-_98457031 | 3.16 |
ENSMUST00000167753.1
|
Gm4450
|
predicted gene 4450 |
| chr1_+_172126750 | 3.11 |
ENSMUST00000075895.2
ENSMUST00000111252.3 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr11_+_78037959 | 3.08 |
ENSMUST00000073660.6
|
Flot2
|
flotillin 2 |
| chr18_+_33464163 | 3.06 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr19_+_56287943 | 3.03 |
ENSMUST00000166049.1
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr12_-_103631404 | 3.03 |
ENSMUST00000121625.1
ENSMUST00000044231.5 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chrX_+_52791179 | 3.01 |
ENSMUST00000101588.1
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr9_+_95559817 | 2.88 |
ENSMUST00000079597.5
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chr19_+_56287911 | 2.87 |
ENSMUST00000095948.4
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr3_-_116712644 | 2.86 |
ENSMUST00000029569.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr7_-_5805445 | 2.77 |
ENSMUST00000075085.6
|
Vmn1r63
|
vomeronasal 1 receptor 63 |
| chr18_+_20310738 | 2.73 |
ENSMUST00000077146.3
|
Dsg1a
|
desmoglein 1 alpha |
| chr6_-_129100903 | 2.72 |
ENSMUST00000032258.6
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr7_+_120173847 | 2.62 |
ENSMUST00000033201.5
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr11_+_46571536 | 2.54 |
ENSMUST00000050937.6
|
BC053393
|
cDNA sequence BC053393 |
| chr8_+_95715901 | 2.53 |
ENSMUST00000034096.4
|
Setd6
|
SET domain containing 6 |
| chr7_-_45062393 | 2.51 |
ENSMUST00000129101.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr9_-_70934808 | 2.46 |
ENSMUST00000034731.8
|
Lipc
|
lipase, hepatic |
| chrM_+_2743 | 2.45 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr4_-_139968026 | 2.40 |
ENSMUST00000105031.2
|
Klhdc7a
|
kelch domain containing 7A |
| chr14_-_36919314 | 2.29 |
ENSMUST00000182797.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr3_+_90537306 | 2.24 |
ENSMUST00000107335.1
|
S100a16
|
S100 calcium binding protein A16 |
| chr11_+_110968016 | 2.24 |
ENSMUST00000106636.1
ENSMUST00000180023.1 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_+_65348386 | 2.21 |
ENSMUST00000031096.7
|
Klb
|
klotho beta |
| chr4_-_150909428 | 2.17 |
ENSMUST00000128075.1
ENSMUST00000105674.1 ENSMUST00000105673.1 |
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr14_-_36919513 | 2.13 |
ENSMUST00000182042.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr5_-_87490869 | 2.10 |
ENSMUST00000147854.1
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr7_-_6011010 | 2.08 |
ENSMUST00000086338.1
|
Vmn1r65
|
vomeronasal 1 receptor 65 |
| chr11_+_118428203 | 2.06 |
ENSMUST00000124861.1
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr11_-_75937726 | 1.97 |
ENSMUST00000121287.1
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr17_+_23660477 | 1.96 |
ENSMUST00000062967.8
|
Ccdc64b
|
coiled-coil domain containing 64B |
| chr7_+_141216626 | 1.96 |
ENSMUST00000141804.1
ENSMUST00000148975.1 |
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr17_+_57062486 | 1.96 |
ENSMUST00000163628.1
|
Crb3
|
crumbs homolog 3 (Drosophila) |
| chr6_+_48647224 | 1.92 |
ENSMUST00000078223.3
|
Gimap8
|
GTPase, IMAP family member 8 |
| chr18_-_66022580 | 1.91 |
ENSMUST00000143990.1
|
Lman1
|
lectin, mannose-binding, 1 |
| chr11_-_75937677 | 1.90 |
ENSMUST00000108420.2
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr6_-_24168083 | 1.87 |
ENSMUST00000031713.8
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr8_-_67974567 | 1.85 |
ENSMUST00000098696.3
ENSMUST00000038959.9 ENSMUST00000093469.4 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_+_78178105 | 1.83 |
ENSMUST00000147819.1
|
Tlcd1
|
TLC domain containing 1 |
| chr3_-_83049797 | 1.82 |
ENSMUST00000048246.3
|
Fgb
|
fibrinogen beta chain |
| chr7_+_83148644 | 1.80 |
ENSMUST00000066005.5
|
A530021J07Rik
|
Riken cDNA A530021J07 gene |
| chr1_+_58795371 | 1.75 |
ENSMUST00000027189.8
|
Casp8
|
caspase 8 |
| chr15_-_10713537 | 1.72 |
ENSMUST00000090339.3
|
Rai14
|
retinoic acid induced 14 |
| chr1_-_187215454 | 1.72 |
ENSMUST00000183819.1
|
Spata17
|
spermatogenesis associated 17 |
| chr14_+_48446128 | 1.72 |
ENSMUST00000124720.1
|
Tmem260
|
transmembrane protein 260 |
| chr1_-_141257158 | 1.62 |
ENSMUST00000179202.1
|
Gm4845
|
predicted gene 4845 |
| chr6_-_128437653 | 1.56 |
ENSMUST00000151796.1
|
Fkbp4
|
FK506 binding protein 4 |
| chr16_-_92358874 | 1.50 |
ENSMUST00000166707.1
|
Kcne1
|
potassium voltage-gated channel, Isk-related subfamily, member 1 |
| chrM_+_7759 | 1.49 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr2_+_91265252 | 1.47 |
ENSMUST00000028691.6
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr14_+_48446340 | 1.44 |
ENSMUST00000111735.2
|
Tmem260
|
transmembrane protein 260 |
| chr13_-_73678005 | 1.39 |
ENSMUST00000022105.7
ENSMUST00000109680.2 ENSMUST00000109679.2 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr2_+_3770673 | 1.35 |
ENSMUST00000177037.1
|
Fam107b
|
family with sequence similarity 107, member B |
| chr19_-_11856001 | 1.33 |
ENSMUST00000079875.3
|
Olfr1418
|
olfactory receptor 1418 |
| chr19_+_26885925 | 1.33 |
ENSMUST00000099536.2
|
Gm815
|
predicted gene 815 |
| chr2_+_125152505 | 1.32 |
ENSMUST00000110494.2
ENSMUST00000028630.2 ENSMUST00000110495.2 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr5_-_106926245 | 1.32 |
ENSMUST00000117588.1
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr6_-_52240841 | 1.32 |
ENSMUST00000121043.1
|
Hoxa10
|
homeobox A10 |
| chr9_+_54980880 | 1.29 |
ENSMUST00000093844.3
|
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr5_-_90640464 | 1.29 |
ENSMUST00000031317.6
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr10_-_7212222 | 1.28 |
ENSMUST00000015346.5
|
Cnksr3
|
Cnksr family member 3 |
| chr2_+_151494274 | 1.27 |
ENSMUST00000028949.9
ENSMUST00000103160.4 |
Nsfl1c
|
NSFL1 (p97) cofactor (p47) |
| chr19_-_30549516 | 1.26 |
ENSMUST00000025803.8
|
Dkk1
|
dickkopf homolog 1 (Xenopus laevis) |
| chr1_-_187215421 | 1.25 |
ENSMUST00000110945.3
ENSMUST00000183931.1 ENSMUST00000027908.6 |
Spata17
|
spermatogenesis associated 17 |
| chr1_-_12991109 | 1.23 |
ENSMUST00000115403.2
ENSMUST00000115402.1 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr17_+_57062231 | 1.23 |
ENSMUST00000097299.3
ENSMUST00000169543.1 ENSMUST00000163763.1 |
Crb3
|
crumbs homolog 3 (Drosophila) |
| chr5_+_95742014 | 1.23 |
ENSMUST00000154086.1
|
Gm7982
|
predicted gene 7982 |
| chrY_-_35130404 | 1.22 |
ENSMUST00000180170.1
|
Gm20855
|
predicted gene, 20855 |
| chr11_+_110968056 | 1.21 |
ENSMUST00000125692.1
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_-_76795489 | 1.15 |
ENSMUST00000082431.3
|
Gpx2
|
glutathione peroxidase 2 |
| chr11_+_23665615 | 1.13 |
ENSMUST00000109525.1
ENSMUST00000020520.4 |
Pus10
|
pseudouridylate synthase 10 |
| chr10_+_79468943 | 1.07 |
ENSMUST00000167976.2
|
Vmn2r83
|
vomeronasal 2, receptor 83 |
| chr8_-_22694061 | 1.04 |
ENSMUST00000131767.1
|
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr19_-_57197556 | 1.00 |
ENSMUST00000099294.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr13_-_21453714 | 0.98 |
ENSMUST00000032820.7
ENSMUST00000110485.1 |
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr5_-_121527186 | 0.98 |
ENSMUST00000152270.1
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr11_+_85832551 | 0.94 |
ENSMUST00000000095.6
|
Tbx2
|
T-box 2 |
| chr8_+_70724064 | 0.91 |
ENSMUST00000034307.7
ENSMUST00000110095.2 |
Pde4c
|
phosphodiesterase 4C, cAMP specific |
| chr9_+_109096659 | 0.91 |
ENSMUST00000130366.1
|
Plxnb1
|
plexin B1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 66.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 5.6 | 27.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 5.5 | 5.5 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 4.4 | 13.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 4.2 | 16.7 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 3.4 | 10.3 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 3.4 | 10.3 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 3.2 | 19.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 3.2 | 9.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 2.8 | 16.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 2.7 | 8.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 2.5 | 17.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 2.5 | 10.0 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 2.3 | 6.9 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 2.3 | 6.9 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 2.1 | 12.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 2.0 | 9.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.9 | 9.6 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.8 | 9.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.7 | 5.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.6 | 8.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 1.6 | 79.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.6 | 7.9 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.6 | 11.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.5 | 24.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 1.4 | 5.7 | GO:0015886 | heme transport(GO:0015886) |
| 1.4 | 5.6 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.3 | 3.9 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 1.3 | 7.6 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 1.2 | 23.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 1.2 | 5.9 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 1.2 | 5.8 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 1.1 | 3.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.1 | 3.3 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 1.1 | 8.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.0 | 24.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 1.0 | 6.0 | GO:0060745 | Sertoli cell proliferation(GO:0060011) prostate epithelial cord elongation(GO:0060523) mammary gland branching involved in pregnancy(GO:0060745) |
| 1.0 | 6.0 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 1.0 | 6.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 1.0 | 3.9 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.9 | 4.6 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.9 | 4.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.8 | 3.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.8 | 2.3 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.7 | 5.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.7 | 3.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.7 | 12.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.6 | 5.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.6 | 1.8 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.6 | 4.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.5 | 4.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.5 | 3.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.5 | 25.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.5 | 3.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 0.5 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.5 | 1.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 4.9 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.4 | 1.3 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.4 | 3.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 0.8 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.4 | 6.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.4 | 3.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 10.3 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.3 | 7.9 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.3 | 3.3 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.3 | 3.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 2.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 0.9 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.3 | 7.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.3 | 3.6 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 | 0.8 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.3 | 1.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.3 | 1.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 30.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.2 | 3.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 2.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 5.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 8.5 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 1.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 8.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 6.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 4.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.4 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.1 | 1.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 12.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.6 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 2.4 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 17.8 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 0.5 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 1.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 1.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 1.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 3.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.4 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 4.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 21.1 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 4.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.9 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 2.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 12.9 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.2 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.0 | 0.6 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0072114 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) pronephros morphogenesis(GO:0072114) mesonephric duct formation(GO:0072181) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 1.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 3.9 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 4.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.0 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of monooxygenase activity(GO:0032769) negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.0 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 3.5 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 2.6 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 2.3 | 9.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 2.1 | 20.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.5 | 4.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 1.4 | 5.5 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.3 | 10.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.0 | 14.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.9 | 6.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.8 | 10.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.8 | 0.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.6 | 1.8 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.6 | 1.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.5 | 3.3 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.5 | 6.9 | GO:0001931 | uropod(GO:0001931) flotillin complex(GO:0016600) cell trailing edge(GO:0031254) |
| 0.5 | 5.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 4.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.3 | 8.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.3 | 42.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 24.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 2.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 4.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 6.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 15.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 6.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.9 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 15.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 4.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 31.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 6.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 17.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 14.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.0 | GO:0035631 | IkappaB kinase complex(GO:0008385) CD40 receptor complex(GO:0035631) |
| 0.1 | 24.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 16.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 9.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 5.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 18.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 41.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 5.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 6.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 5.6 | 16.8 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 4.4 | 17.7 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 4.2 | 12.6 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 3.9 | 19.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 3.8 | 19.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 3.8 | 11.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 3.7 | 47.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 3.6 | 14.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 3.5 | 24.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 3.4 | 10.3 | GO:0070540 | stearic acid binding(GO:0070540) |
| 3.3 | 13.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 3.1 | 117.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 2.5 | 10.0 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 2.3 | 6.9 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 2.1 | 8.5 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 2.0 | 6.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.9 | 13.6 | GO:0005534 | galactose binding(GO:0005534) |
| 1.6 | 9.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.4 | 5.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.1 | 5.6 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 1.1 | 7.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 1.1 | 3.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 1.1 | 3.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.0 | 16.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 1.0 | 6.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.0 | 22.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 1.0 | 4.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.9 | 4.7 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.9 | 10.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.9 | 3.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.7 | 2.2 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.7 | 9.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.7 | 4.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.6 | 4.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 12.4 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.6 | 5.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 4.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.5 | 11.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.5 | 12.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 1.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 1.8 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.4 | 2.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 6.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.3 | 8.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.3 | 3.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.3 | 3.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 0.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.3 | 4.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 5.5 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.3 | 3.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 3.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 23.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.2 | 6.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 5.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 3.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 1.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 0.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 5.1 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.2 | 7.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 0.8 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 10.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 13.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 5.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 7.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 3.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 6.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 3.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 3.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 12.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 2.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 3.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 6.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 11.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 11.7 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 12.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 6.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 1.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 5.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 4.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 5.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.8 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 19.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 9.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 15.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 4.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 8.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 1.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 6.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 6.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 7.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 10.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 7.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 17.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 12.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 32.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 3.2 | 41.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.7 | 13.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.4 | 18.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 1.3 | 18.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.0 | 19.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.9 | 23.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.9 | 17.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.8 | 23.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.7 | 9.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.7 | 10.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.5 | 10.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 3.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 21.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.4 | 10.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 20.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 3.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 9.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 7.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 5.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 4.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 4.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 8.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 8.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 1.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 6.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 2.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 2.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 3.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 27.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 12.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 7.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 5.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 6.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 6.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 5.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 3.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |