Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
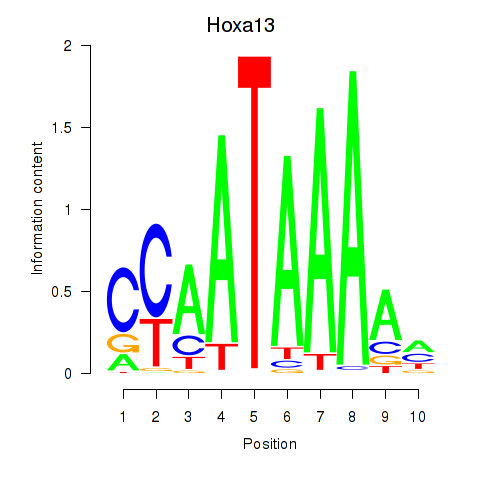
Results for Hoxa13
Z-value: 1.78
Transcription factors associated with Hoxa13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa13
|
ENSMUSG00000038203.12 | homeobox A13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa13 | mm10_v2_chr6_-_52260822_52260880 | -0.64 | 2.7e-05 | Click! |
Activity profile of Hoxa13 motif
Sorted Z-values of Hoxa13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_61674094 | 26.99 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_61303802 | 23.50 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr4_-_61303998 | 22.16 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_61439743 | 18.15 |
ENSMUST00000095049.4
|
Mup15
|
major urinary protein 15 |
| chr4_-_61519467 | 17.91 |
ENSMUST00000095051.5
ENSMUST00000107483.1 |
Mup16
|
major urinary protein 16 |
| chr4_-_61228271 | 17.78 |
ENSMUST00000072678.5
ENSMUST00000098042.3 |
Mup13
|
major urinary protein 13 |
| chr4_-_60070411 | 17.46 |
ENSMUST00000079697.3
ENSMUST00000125282.1 ENSMUST00000166098.1 |
Mup7
|
major urinary protein 7 |
| chr4_-_60662358 | 16.46 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr4_-_61595871 | 16.41 |
ENSMUST00000107484.1
|
Mup17
|
major urinary protein 17 |
| chr4_-_60222580 | 16.34 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_60582152 | 15.86 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr4_-_60139857 | 15.37 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr4_-_60501903 | 15.34 |
ENSMUST00000084548.4
ENSMUST00000103012.3 ENSMUST00000107499.3 |
Mup1
|
major urinary protein 1 |
| chr4_-_60421933 | 14.77 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_60741275 | 14.68 |
ENSMUST00000117932.1
|
Mup12
|
major urinary protein 12 |
| chr7_-_30924169 | 11.43 |
ENSMUST00000074671.6
|
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr19_+_40089688 | 11.01 |
ENSMUST00000068094.6
ENSMUST00000080171.2 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr8_-_93279717 | 10.93 |
ENSMUST00000034178.8
|
Ces1f
|
carboxylesterase 1F |
| chr8_-_24576297 | 10.30 |
ENSMUST00000033953.7
ENSMUST00000121992.1 |
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr8_-_5105232 | 10.17 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr4_+_63362443 | 8.73 |
ENSMUST00000075341.3
|
Orm2
|
orosomucoid 2 |
| chr15_+_6445320 | 7.64 |
ENSMUST00000022749.9
|
C9
|
complement component 9 |
| chr1_-_150465563 | 6.74 |
ENSMUST00000164600.1
ENSMUST00000111902.2 ENSMUST00000111901.2 ENSMUST00000006171.9 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr11_-_113710017 | 6.68 |
ENSMUST00000018871.1
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr12_-_103657095 | 5.22 |
ENSMUST00000152517.1
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr10_-_116896879 | 4.76 |
ENSMUST00000048229.7
|
Myrfl
|
myelin regulatory factor-like |
| chr8_+_45069137 | 4.65 |
ENSMUST00000067984.7
|
Mtnr1a
|
melatonin receptor 1A |
| chr1_+_59256906 | 4.50 |
ENSMUST00000160662.1
ENSMUST00000114248.2 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr16_+_56204313 | 4.37 |
ENSMUST00000160116.1
ENSMUST00000069936.7 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr5_-_87337165 | 4.27 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr7_+_123462274 | 4.24 |
ENSMUST00000033023.3
|
Aqp8
|
aquaporin 8 |
| chr19_+_24875679 | 4.22 |
ENSMUST00000073080.5
|
Gm10053
|
predicted gene 10053 |
| chr1_-_97128249 | 4.12 |
ENSMUST00000027569.7
|
Slco6c1
|
solute carrier organic anion transporter family, member 6c1 |
| chr19_-_6067785 | 4.10 |
ENSMUST00000162575.1
ENSMUST00000159084.1 ENSMUST00000161718.1 ENSMUST00000162810.1 ENSMUST00000025713.5 ENSMUST00000113543.2 ENSMUST00000161528.1 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr12_-_103657159 | 4.07 |
ENSMUST00000044159.6
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr7_-_30944017 | 4.04 |
ENSMUST00000062620.7
|
Hamp
|
hepcidin antimicrobial peptide |
| chr4_+_144893127 | 3.99 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr10_-_92375367 | 3.99 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr10_-_115362191 | 3.85 |
ENSMUST00000092170.5
|
Tmem19
|
transmembrane protein 19 |
| chr11_+_76904475 | 3.51 |
ENSMUST00000142166.1
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr8_+_45069374 | 3.47 |
ENSMUST00000130141.1
|
Mtnr1a
|
melatonin receptor 1A |
| chr12_-_104153846 | 3.44 |
ENSMUST00000085050.3
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr16_+_11405648 | 3.40 |
ENSMUST00000096273.2
|
Snx29
|
sorting nexin 29 |
| chr1_-_121332545 | 3.31 |
ENSMUST00000161068.1
|
Insig2
|
insulin induced gene 2 |
| chr17_+_31433054 | 3.25 |
ENSMUST00000136384.1
|
Pde9a
|
phosphodiesterase 9A |
| chr9_+_7445822 | 3.20 |
ENSMUST00000034497.6
|
Mmp3
|
matrix metallopeptidase 3 |
| chr19_+_12633507 | 3.19 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr1_-_121332571 | 3.06 |
ENSMUST00000071064.6
|
Insig2
|
insulin induced gene 2 |
| chr11_-_11898092 | 3.05 |
ENSMUST00000178704.1
|
Ddc
|
dopa decarboxylase |
| chr13_-_23934156 | 3.03 |
ENSMUST00000052776.2
|
Hist1h2ba
|
histone cluster 1, H2ba |
| chr5_-_28467093 | 3.02 |
ENSMUST00000002708.3
|
Shh
|
sonic hedgehog |
| chr13_-_23914998 | 3.01 |
ENSMUST00000021769.8
ENSMUST00000110407.2 |
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr8_+_45507768 | 2.98 |
ENSMUST00000067065.7
ENSMUST00000098788.3 ENSMUST00000067107.7 ENSMUST00000171337.2 ENSMUST00000138049.1 ENSMUST00000141039.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_+_138313279 | 2.95 |
ENSMUST00000013455.6
ENSMUST00000106247.1 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr9_-_76323559 | 2.93 |
ENSMUST00000063140.8
|
Hcrtr2
|
hypocretin (orexin) receptor 2 |
| chr11_-_73324616 | 2.88 |
ENSMUST00000021119.2
|
Aspa
|
aspartoacylase |
| chr3_+_96219858 | 2.86 |
ENSMUST00000073115.4
|
Hist2h2ab
|
histone cluster 2, H2ab |
| chr15_+_62178175 | 2.85 |
ENSMUST00000182476.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr3_-_10331358 | 2.81 |
ENSMUST00000065938.8
ENSMUST00000118410.1 |
Impa1
|
inositol (myo)-1(or 4)-monophosphatase 1 |
| chr19_+_12633303 | 2.76 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr9_-_44799179 | 2.71 |
ENSMUST00000114705.1
ENSMUST00000002100.7 |
Tmem25
|
transmembrane protein 25 |
| chr8_+_95352258 | 2.70 |
ENSMUST00000034243.5
|
Mmp15
|
matrix metallopeptidase 15 |
| chr1_-_193264006 | 2.69 |
ENSMUST00000161737.1
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr6_+_40628824 | 2.65 |
ENSMUST00000071535.6
|
Mgam
|
maltase-glucoamylase |
| chrX_-_162565514 | 2.63 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr4_-_134853294 | 2.61 |
ENSMUST00000030628.8
|
Tmem57
|
transmembrane protein 57 |
| chr17_-_25844417 | 2.55 |
ENSMUST00000176591.1
|
Rhot2
|
ras homolog gene family, member T2 |
| chr4_+_148140699 | 2.52 |
ENSMUST00000140049.1
ENSMUST00000105707.1 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr14_-_61556881 | 2.52 |
ENSMUST00000022497.8
|
Spryd7
|
SPRY domain containing 7 |
| chr1_-_72212249 | 2.51 |
ENSMUST00000048860.7
|
Mreg
|
melanoregulin |
| chr10_-_109010955 | 2.49 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr16_-_23988852 | 2.49 |
ENSMUST00000023151.5
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr14_-_68533689 | 2.49 |
ENSMUST00000022640.7
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr13_-_71963713 | 2.48 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr14_+_65969714 | 2.44 |
ENSMUST00000153460.1
|
Clu
|
clusterin |
| chr13_+_34734837 | 2.44 |
ENSMUST00000039605.6
|
Fam50b
|
family with sequence similarity 50, member B |
| chr6_+_3993776 | 2.42 |
ENSMUST00000031673.5
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chrM_+_3906 | 2.38 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr2_-_18048784 | 2.33 |
ENSMUST00000142856.1
|
Skida1
|
SKI/DACH domain containing 1 |
| chr18_+_38418946 | 2.32 |
ENSMUST00000025293.3
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr18_-_3281752 | 2.32 |
ENSMUST00000140332.1
ENSMUST00000147138.1 |
Crem
|
cAMP responsive element modulator |
| chr7_+_43690418 | 2.31 |
ENSMUST00000056329.6
|
Klk14
|
kallikrein related-peptidase 14 |
| chr1_-_36244245 | 2.29 |
ENSMUST00000046875.7
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr2_+_126215100 | 2.28 |
ENSMUST00000164042.2
|
Gm17555
|
predicted gene, 17555 |
| chr2_+_162987330 | 2.26 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_+_162987502 | 2.24 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr7_-_73537621 | 2.24 |
ENSMUST00000172704.1
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr15_-_58214882 | 2.22 |
ENSMUST00000022986.6
|
Fbxo32
|
F-box protein 32 |
| chr8_+_4349588 | 2.20 |
ENSMUST00000110982.1
ENSMUST00000024004.7 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr2_-_91195035 | 2.19 |
ENSMUST00000111356.1
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_+_164503306 | 2.19 |
ENSMUST00000181831.1
|
Gm26685
|
predicted gene, 26685 |
| chr5_+_146079254 | 2.18 |
ENSMUST00000035571.8
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr12_+_59129757 | 2.15 |
ENSMUST00000069430.8
ENSMUST00000177370.1 |
Ctage5
|
CTAGE family, member 5 |
| chr13_+_23684192 | 2.13 |
ENSMUST00000018246.4
|
Hist1h2bc
|
histone cluster 1, H2bc |
| chr2_-_91195097 | 2.13 |
ENSMUST00000002177.2
ENSMUST00000111354.1 |
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr9_+_43829963 | 2.12 |
ENSMUST00000180221.1
|
Gm3898
|
predicted gene 3898 |
| chr7_+_103550368 | 2.11 |
ENSMUST00000106888.1
|
Olfr613
|
olfactory receptor 613 |
| chr6_+_79818031 | 2.07 |
ENSMUST00000179797.1
|
Gm20594
|
predicted gene, 20594 |
| chr5_-_87538188 | 2.01 |
ENSMUST00000031199.4
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr3_-_28765364 | 2.00 |
ENSMUST00000094335.3
|
Gm6505
|
predicted pseudogene 6505 |
| chr9_+_45319072 | 2.00 |
ENSMUST00000034597.7
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr8_+_129118043 | 1.99 |
ENSMUST00000108744.1
|
1700008F21Rik
|
RIKEN cDNA 1700008F21 gene |
| chr1_+_107361929 | 1.96 |
ENSMUST00000027566.2
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr15_+_3270767 | 1.94 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr12_+_59129720 | 1.92 |
ENSMUST00000175912.1
ENSMUST00000176892.1 |
Ctage5
|
CTAGE family, member 5 |
| chr10_+_59395632 | 1.88 |
ENSMUST00000092511.4
|
Gm10273
|
predicted pseudogene 10273 |
| chr10_-_68278713 | 1.86 |
ENSMUST00000020106.7
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr13_+_4574075 | 1.85 |
ENSMUST00000021628.3
|
Akr1c21
|
aldo-keto reductase family 1, member C21 |
| chr1_-_175625580 | 1.85 |
ENSMUST00000027810.7
|
Fh1
|
fumarate hydratase 1 |
| chr19_+_46707443 | 1.84 |
ENSMUST00000003655.7
|
As3mt
|
arsenic (+3 oxidation state) methyltransferase |
| chr11_-_17211504 | 1.83 |
ENSMUST00000020317.7
|
Pno1
|
partner of NOB1 homolog (S. cerevisiae) |
| chrX_+_139563316 | 1.82 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr14_-_47189406 | 1.81 |
ENSMUST00000089959.6
|
Gch1
|
GTP cyclohydrolase 1 |
| chr2_-_91194767 | 1.80 |
ENSMUST00000111355.1
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_-_19023538 | 1.79 |
ENSMUST00000036018.5
|
Foxa3
|
forkhead box A3 |
| chrM_-_14060 | 1.78 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_+_99030946 | 1.76 |
ENSMUST00000030280.6
|
Angptl3
|
angiopoietin-like 3 |
| chr4_+_41762309 | 1.74 |
ENSMUST00000108042.2
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr15_-_76232045 | 1.72 |
ENSMUST00000167754.1
|
Plec
|
plectin |
| chr7_+_127800604 | 1.70 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_+_185332143 | 1.69 |
ENSMUST00000027916.6
ENSMUST00000151769.1 ENSMUST00000110965.1 |
Bpnt1
|
bisphosphate 3'-nucleotidase 1 |
| chr3_+_114904062 | 1.68 |
ENSMUST00000081752.6
|
Olfm3
|
olfactomedin 3 |
| chr9_-_51328898 | 1.67 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr4_+_152199805 | 1.66 |
ENSMUST00000105652.2
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr1_+_175632169 | 1.65 |
ENSMUST00000097458.3
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_-_144550777 | 1.64 |
ENSMUST00000028915.5
|
Rbbp9
|
retinoblastoma binding protein 9 |
| chr4_+_89137122 | 1.63 |
ENSMUST00000058030.7
|
Mtap
|
methylthioadenosine phosphorylase |
| chr1_+_175631996 | 1.62 |
ENSMUST00000040250.8
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr6_-_125165707 | 1.61 |
ENSMUST00000118875.1
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr7_-_141434532 | 1.59 |
ENSMUST00000133021.1
ENSMUST00000106007.3 ENSMUST00000150026.1 ENSMUST00000133206.2 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr19_+_26623419 | 1.57 |
ENSMUST00000176584.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_48559971 | 1.56 |
ENSMUST00000169406.1
|
Gm1818
|
predicted gene 1818 |
| chr15_-_75431745 | 1.56 |
ENSMUST00000096397.1
|
9030619P08Rik
|
RIKEN cDNA 9030619P08 gene |
| chr2_-_38926217 | 1.55 |
ENSMUST00000076275.4
ENSMUST00000142130.1 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr10_+_28668560 | 1.55 |
ENSMUST00000161345.1
|
Themis
|
thymocyte selection associated |
| chr13_-_92131494 | 1.52 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr1_-_162866502 | 1.52 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr7_+_5015466 | 1.51 |
ENSMUST00000086349.3
|
Zfp524
|
zinc finger protein 524 |
| chr18_-_61036189 | 1.49 |
ENSMUST00000025521.8
|
Cdx1
|
caudal type homeobox 1 |
| chr19_-_5796924 | 1.49 |
ENSMUST00000174808.1
|
Malat1
|
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
| chr3_+_66985647 | 1.48 |
ENSMUST00000162362.1
ENSMUST00000065074.7 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr13_+_119606650 | 1.47 |
ENSMUST00000178948.1
|
Gm21967
|
predicted gene, 21967 |
| chr11_+_96194299 | 1.46 |
ENSMUST00000062709.3
|
Hoxb13
|
homeobox B13 |
| chr17_-_25844514 | 1.45 |
ENSMUST00000176709.1
|
Rhot2
|
ras homolog gene family, member T2 |
| chr2_-_89024099 | 1.44 |
ENSMUST00000099799.2
|
Olfr1217
|
olfactory receptor 1217 |
| chr4_-_42661893 | 1.43 |
ENSMUST00000108006.3
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr10_-_34044302 | 1.43 |
ENSMUST00000048052.5
|
Fam26d
|
family with sequence similarity 26, member D |
| chr9_-_21067093 | 1.43 |
ENSMUST00000115494.2
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr3_-_89411781 | 1.42 |
ENSMUST00000107429.3
ENSMUST00000129308.2 ENSMUST00000107426.1 ENSMUST00000050398.4 ENSMUST00000162701.1 |
Flad1
|
RFad1, flavin adenine dinucleotide synthetase, homolog (yeast) |
| chr10_+_24076500 | 1.41 |
ENSMUST00000051133.5
|
Taar8a
|
trace amine-associated receptor 8A |
| chr13_-_91388079 | 1.40 |
ENSMUST00000181054.1
|
A830009L08Rik
|
RIKEN cDNA A830009L08 gene |
| chr19_-_11081088 | 1.39 |
ENSMUST00000025636.6
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr8_-_25038875 | 1.38 |
ENSMUST00000084031.4
|
Htra4
|
HtrA serine peptidase 4 |
| chr12_-_86726439 | 1.38 |
ENSMUST00000021682.8
|
Angel1
|
angel homolog 1 (Drosophila) |
| chr14_+_55971428 | 1.37 |
ENSMUST00000089555.2
|
Cma2
|
chymase 2, mast cell |
| chr5_-_62766153 | 1.37 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_+_40540751 | 1.36 |
ENSMUST00000091186.3
|
Intu
|
inturned planar cell polarity effector homolog (Drosophila) |
| chr13_+_32802007 | 1.36 |
ENSMUST00000021832.6
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr11_+_76902152 | 1.36 |
ENSMUST00000102495.1
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr17_-_35897073 | 1.35 |
ENSMUST00000150056.1
ENSMUST00000156817.1 ENSMUST00000146451.1 ENSMUST00000148482.1 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr12_+_108792946 | 1.34 |
ENSMUST00000021692.7
|
Yy1
|
YY1 transcription factor |
| chr7_+_34912379 | 1.34 |
ENSMUST00000075068.7
|
Pepd
|
peptidase D |
| chr13_+_23934434 | 1.33 |
ENSMUST00000072391.1
|
Hist1h2aa
|
histone cluster 1, H2aa |
| chr9_+_107340593 | 1.33 |
ENSMUST00000042581.2
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr15_+_25773985 | 1.33 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr6_-_24528013 | 1.33 |
ENSMUST00000023851.5
|
Ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr13_-_107414742 | 1.31 |
ENSMUST00000061241.6
|
Apoo-ps
|
apolipoprotein O, pseudogene |
| chr3_+_14480692 | 1.31 |
ENSMUST00000037321.7
ENSMUST00000120484.1 ENSMUST00000120801.1 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr7_-_41499890 | 1.31 |
ENSMUST00000098509.3
|
AW146154
|
expressed sequence AW146154 |
| chr6_+_129180613 | 1.30 |
ENSMUST00000032260.5
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr6_-_125165576 | 1.30 |
ENSMUST00000183272.1
ENSMUST00000182052.1 ENSMUST00000182277.1 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_135284075 | 1.30 |
ENSMUST00000077340.7
ENSMUST00000074357.7 |
Rnpep
|
arginyl aminopeptidase (aminopeptidase B) |
| chr16_+_29579347 | 1.29 |
ENSMUST00000038867.6
ENSMUST00000161186.1 |
Opa1
|
optic atrophy 1 |
| chr12_-_72085393 | 1.29 |
ENSMUST00000019862.2
|
L3hypdh
|
L-3-hydroxyproline dehydratase (trans-) |
| chr17_+_25829036 | 1.29 |
ENSMUST00000026832.7
ENSMUST00000123582.1 |
Jmjd8
|
jumonji domain containing 8 |
| chr1_+_87594545 | 1.28 |
ENSMUST00000165109.1
ENSMUST00000070898.5 |
Neu2
|
neuraminidase 2 |
| chr10_+_116301374 | 1.27 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr6_-_38354243 | 1.26 |
ENSMUST00000114900.1
|
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr7_+_44198191 | 1.25 |
ENSMUST00000085450.2
|
Klk1b3
|
kallikrein 1-related peptidase b3 |
| chr2_-_140066661 | 1.25 |
ENSMUST00000046656.2
ENSMUST00000099304.3 ENSMUST00000110079.2 |
Tasp1
|
taspase, threonine aspartase 1 |
| chr16_+_34649920 | 1.25 |
ENSMUST00000148562.1
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr3_+_106034661 | 1.25 |
ENSMUST00000170669.2
|
Gm4540
|
predicted gene 4540 |
| chr5_-_93671366 | 1.24 |
ENSMUST00000160418.1
ENSMUST00000159578.1 ENSMUST00000162539.1 ENSMUST00000160382.1 ENSMUST00000076321.6 ENSMUST00000101024.3 ENSMUST00000159489.1 ENSMUST00000162964.1 |
C87414
|
expressed sequence C87414 |
| chr5_-_24842579 | 1.23 |
ENSMUST00000030787.8
|
Rheb
|
Ras homolog enriched in brain |
| chr17_-_35897371 | 1.23 |
ENSMUST00000148721.1
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr19_-_43674844 | 1.21 |
ENSMUST00000046038.7
|
Slc25a28
|
solute carrier family 25, member 28 |
| chr11_+_97029925 | 1.20 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr11_+_83473079 | 1.20 |
ENSMUST00000021018.4
|
Taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_+_4434306 | 1.20 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr2_+_32395896 | 1.19 |
ENSMUST00000028162.3
|
Ptges2
|
prostaglandin E synthase 2 |
| chr11_+_98446826 | 1.18 |
ENSMUST00000019456.4
|
Grb7
|
growth factor receptor bound protein 7 |
| chr7_+_43430088 | 1.17 |
ENSMUST00000004732.5
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr2_+_3424123 | 1.17 |
ENSMUST00000061852.5
ENSMUST00000100463.3 ENSMUST00000102988.3 ENSMUST00000115066.1 |
Dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr5_-_121527186 | 1.16 |
ENSMUST00000152270.1
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr7_-_135528645 | 1.15 |
ENSMUST00000053716.7
|
Clrn3
|
clarin 3 |
| chr15_+_31224371 | 1.14 |
ENSMUST00000044524.9
|
Dap
|
death-associated protein |
| chr19_+_12674179 | 1.14 |
ENSMUST00000057924.2
|
Olfr1442
|
olfactory receptor 1442 |
| chr8_-_41054771 | 1.14 |
ENSMUST00000093534.4
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_63184253 | 1.13 |
ENSMUST00000075052.3
ENSMUST00000112454.1 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr16_-_31314804 | 1.12 |
ENSMUST00000115230.1
ENSMUST00000130560.1 |
Apod
|
apolipoprotein D |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.5 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 3.6 | 10.9 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.9 | 31.8 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 2.0 | 6.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.2 | 13.6 | GO:0070189 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 1.2 | 3.7 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 1.2 | 4.8 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.0 | 11.0 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.0 | 2.9 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.8 | 6.7 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.7 | 2.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 2.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.7 | 4.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.6 | 2.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.6 | 2.5 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.6 | 1.8 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.6 | 6.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.6 | 2.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.5 | 2.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.5 | 10.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 2.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.5 | 7.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 1.4 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.5 | 8.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.5 | 1.8 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.4 | 1.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.4 | 1.8 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.4 | 3.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.4 | 1.7 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.4 | 1.6 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.4 | 2.4 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.4 | 1.1 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 1.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.4 | 9.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.4 | 1.1 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.4 | 5.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 0.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.4 | 1.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 3.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 2.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 0.7 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.3 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 1.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 0.9 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.3 | 4.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 0.8 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.3 | 2.5 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.3 | 1.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 4.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 1.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.2 | 0.7 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 1.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 2.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 2.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.9 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.2 | 1.3 | GO:0014042 | positive regulation of mitochondrial fusion(GO:0010636) positive regulation of neuron maturation(GO:0014042) |
| 0.2 | 1.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 0.6 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 0.6 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 1.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 4.0 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 3.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.8 | GO:0060296 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 1.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.2 | 1.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.2 | 0.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 1.3 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 2.3 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 1.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 1.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 1.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 3.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 2.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 0.6 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 1.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 1.3 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 0.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.6 | GO:0061043 | positive regulation of vascular wound healing(GO:0035470) regulation of vascular wound healing(GO:0061043) |
| 0.1 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.6 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 2.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 1.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 1.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 1.0 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.4 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 3.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 1.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 4.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 6.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.2 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 | 1.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.7 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 2.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 2.6 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 3.1 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 1.0 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.3 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.7 | GO:0051036 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 2.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.7 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.3 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 1.5 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 2.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.4 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.1 | 1.0 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 2.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 2.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.2 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.2 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.6 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.3 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 0.5 | GO:0060068 | vagina development(GO:0060068) |
| 0.1 | 0.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.1 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 1.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 2.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 2.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 2.8 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 2.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.9 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.4 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.6 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.5 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.0 | 1.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 1.4 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 1.4 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 1.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 3.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 1.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.9 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 1.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 5.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.8 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 1.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 1.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.4 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.0 | 3.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.7 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.8 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.4 | 5.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 1.4 | 4.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.8 | 6.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.8 | 0.8 | GO:0071920 | cleavage body(GO:0071920) |
| 0.4 | 1.8 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.4 | 2.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 2.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 1.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 6.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 1.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 1.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.7 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 1.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 0.9 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 2.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 4.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 4.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 11.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 9.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 8.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 3.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 2.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 2.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 15.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 6.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 2.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 3.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 8.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 3.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 3.0 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 27.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.3 | GO:0005009 | insulin-activated receptor activity(GO:0005009) pheromone activity(GO:0005186) |
| 3.4 | 10.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 2.8 | 11.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.7 | 8.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.5 | 5.9 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.5 | 10.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.2 | 6.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.0 | 2.9 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 1.0 | 2.9 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.9 | 2.8 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.9 | 2.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.8 | 3.1 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.7 | 4.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.7 | 3.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.6 | 2.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.6 | 1.9 | GO:0070401 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.6 | 14.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.6 | 2.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.5 | 3.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.5 | 1.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.5 | 2.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.5 | 3.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 1.7 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.4 | 1.3 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.4 | 1.7 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.4 | 2.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 1.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 1.1 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 1.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 1.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 4.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 7.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.3 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 0.8 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 1.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 1.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 0.7 | GO:0004794 | L-serine ammonia-lyase activity(GO:0003941) L-threonine ammonia-lyase activity(GO:0004794) |
| 0.2 | 0.7 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 6.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 3.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 2.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 2.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.7 | GO:0030977 | taurine binding(GO:0030977) |
| 0.2 | 0.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 0.6 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.2 | 0.9 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 5.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 1.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 1.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.8 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.2 | 2.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 4.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 0.7 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 0.4 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.2 | 2.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 1.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 2.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 6.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.7 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 1.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.8 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 4.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 1.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 4.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 2.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 2.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.9 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 2.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 4.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 10.4 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 9.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.0 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 2.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 11.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 7.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 1.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.0 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 3.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.5 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 2.6 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 6.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 6.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 4.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.9 | 2.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 13.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 2.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.4 | 4.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 3.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 7.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 2.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 5.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 4.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 5.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 6.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 8.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 2.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 3.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.5 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 2.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 5.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |