Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
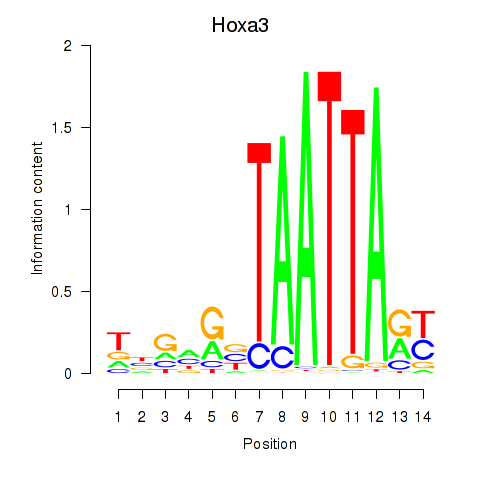
Results for Hoxa3
Z-value: 0.26
Transcription factors associated with Hoxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa3
|
ENSMUSG00000079560.7 | homeobox A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa3 | mm10_v2_chr6_-_52183828_52183855 | 0.14 | 4.1e-01 | Click! |
Activity profile of Hoxa3 motif
Sorted Z-values of Hoxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7966000 | 0.88 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr6_-_136781718 | 0.64 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr14_+_27000362 | 0.63 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr2_-_168767136 | 0.59 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr2_-_168767029 | 0.45 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr2_+_173153048 | 0.39 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr9_+_119063429 | 0.33 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr16_-_42340595 | 0.30 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr10_-_37138863 | 0.30 |
ENSMUST00000092584.5
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr15_-_34356421 | 0.26 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr10_+_37139558 | 0.23 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr2_-_164171113 | 0.18 |
ENSMUST00000045196.3
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr2_+_36230426 | 0.18 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_-_86548268 | 0.16 |
ENSMUST00000077524.3
|
Mab21l2
|
mab-21-like 2 (C. elegans) |
| chr7_-_38019505 | 0.14 |
ENSMUST00000085513.4
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr2_-_116067391 | 0.13 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr3_+_159839729 | 0.12 |
ENSMUST00000068952.5
|
Wls
|
wntless homolog (Drosophila) |
| chr1_-_172027269 | 0.11 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr19_-_55241236 | 0.11 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr12_+_59013379 | 0.11 |
ENSMUST00000021379.7
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr5_+_90561102 | 0.10 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr3_+_55782500 | 0.10 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr7_-_45103747 | 0.09 |
ENSMUST00000003512.7
|
Fcgrt
|
Fc receptor, IgG, alpha chain transporter |
| chr14_+_79515618 | 0.08 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr6_+_63255971 | 0.08 |
ENSMUST00000159561.1
ENSMUST00000095852.3 |
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr10_+_40349265 | 0.08 |
ENSMUST00000044672.4
ENSMUST00000095743.2 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr10_+_26772477 | 0.07 |
ENSMUST00000039557.7
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr4_-_14621805 | 0.07 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_-_137531952 | 0.07 |
ENSMUST00000140139.1
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr17_+_17402672 | 0.06 |
ENSMUST00000115576.2
|
Lix1
|
limb expression 1 homolog (chicken) |
| chr17_-_70853482 | 0.06 |
ENSMUST00000118283.1
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr5_+_138187485 | 0.05 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr11_-_99374895 | 0.05 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr16_-_29544852 | 0.04 |
ENSMUST00000039090.8
|
Atp13a4
|
ATPase type 13A4 |
| chr9_-_77399308 | 0.04 |
ENSMUST00000183878.1
|
RP23-264N13.2
|
RP23-264N13.2 |
| chr7_-_116038734 | 0.04 |
ENSMUST00000166877.1
|
Sox6
|
SRY-box containing gene 6 |
| chr5_+_104202609 | 0.03 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr12_+_71170589 | 0.03 |
ENSMUST00000129376.1
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr11_+_95384662 | 0.02 |
ENSMUST00000021243.7
ENSMUST00000146556.1 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr4_+_108719649 | 0.02 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr16_-_92400067 | 0.02 |
ENSMUST00000023672.8
|
Rcan1
|
regulator of calcineurin 1 |
| chr5_-_62766153 | 0.01 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_+_116067213 | 0.01 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chrX_+_16619698 | 0.01 |
ENSMUST00000026013.5
|
Maoa
|
monoamine oxidase A |
| chr5_-_137531204 | 0.01 |
ENSMUST00000150063.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr14_+_64589802 | 0.00 |
ENSMUST00000180610.1
|
A930011O12Rik
|
RIKEN cDNA A930011O12 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.1 | 0.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |