Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
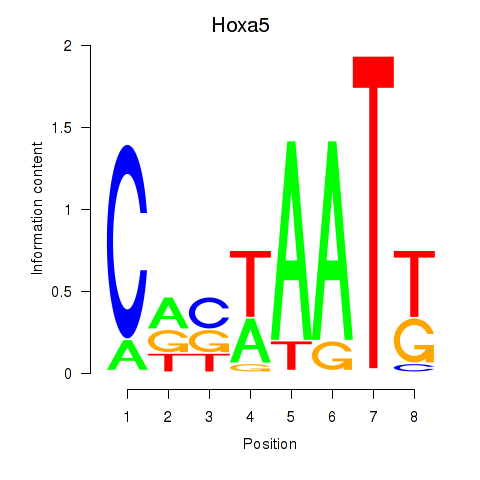
Results for Hoxa5
Z-value: 1.03
Transcription factors associated with Hoxa5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa5
|
ENSMUSG00000038253.6 | homeobox A5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa5 | mm10_v2_chr6_-_52204415_52204587 | 0.28 | 9.2e-02 | Click! |
Activity profile of Hoxa5 motif
Sorted Z-values of Hoxa5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142661858 | 3.30 |
ENSMUST00000145896.2
|
Igf2
|
insulin-like growth factor 2 |
| chr16_+_32756336 | 2.94 |
ENSMUST00000135753.1
|
Muc4
|
mucin 4 |
| chr14_-_47418407 | 2.05 |
ENSMUST00000043296.3
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr7_+_26061495 | 2.04 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr3_+_51661167 | 2.03 |
ENSMUST00000099106.3
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr11_-_83286722 | 2.00 |
ENSMUST00000163961.2
|
Slfn14
|
schlafen family member 14 |
| chr3_-_98893209 | 1.92 |
ENSMUST00000029464.7
|
Hao2
|
hydroxyacid oxidase 2 |
| chr8_-_85380964 | 1.88 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr13_+_23535411 | 1.72 |
ENSMUST00000080859.5
|
Hist1h3g
|
histone cluster 1, H3g |
| chr6_+_86078070 | 1.69 |
ENSMUST00000032069.5
|
Add2
|
adducin 2 (beta) |
| chr9_-_36726374 | 1.68 |
ENSMUST00000172702.2
ENSMUST00000172742.1 ENSMUST00000034625.5 |
Chek1
|
checkpoint kinase 1 |
| chr17_+_21691860 | 1.64 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
| chr7_-_14254870 | 1.63 |
ENSMUST00000184731.1
ENSMUST00000076576.6 |
Sult2a6
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6 |
| chr7_-_142661305 | 1.58 |
ENSMUST00000105936.1
|
Igf2
|
insulin-like growth factor 2 |
| chr13_+_104229366 | 1.54 |
ENSMUST00000022227.6
|
Cenpk
|
centromere protein K |
| chr3_-_7613427 | 1.47 |
ENSMUST00000168269.2
|
Il7
|
interleukin 7 |
| chr4_-_46413486 | 1.46 |
ENSMUST00000071096.2
|
Hemgn
|
hemogen |
| chr6_-_49214954 | 1.41 |
ENSMUST00000031838.7
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr7_-_14562171 | 1.38 |
ENSMUST00000181796.1
|
Vmn1r90
|
vomeronasal 1 receptor 90 |
| chr14_-_56102458 | 1.37 |
ENSMUST00000015583.1
|
Ctsg
|
cathepsin G |
| chr7_-_135716374 | 1.37 |
ENSMUST00000033310.7
|
Mki67
|
antigen identified by monoclonal antibody Ki 67 |
| chr2_-_13491900 | 1.36 |
ENSMUST00000091436.5
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr1_-_169531343 | 1.33 |
ENSMUST00000028000.7
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr15_-_34356421 | 1.32 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr8_+_21655780 | 1.31 |
ENSMUST00000079528.5
|
Defa17
|
defensin, alpha, 17 |
| chr3_+_28781305 | 1.31 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr8_+_21287409 | 1.29 |
ENSMUST00000098893.3
|
Defa3
|
defensin, alpha, 3 |
| chr6_-_60829826 | 1.28 |
ENSMUST00000163779.1
|
Snca
|
synuclein, alpha |
| chr4_-_119189949 | 1.27 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr2_+_84734050 | 1.25 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr4_+_121039385 | 1.23 |
ENSMUST00000030372.5
|
Col9a2
|
collagen, type IX, alpha 2 |
| chr2_-_28621932 | 1.20 |
ENSMUST00000028156.7
ENSMUST00000164290.1 |
Gfi1b
|
growth factor independent 1B |
| chr14_-_52036143 | 1.19 |
ENSMUST00000052560.4
|
Olfr221
|
olfactory receptor 221 |
| chr14_+_65806066 | 1.18 |
ENSMUST00000139644.1
|
Pbk
|
PDZ binding kinase |
| chr13_-_49652714 | 1.15 |
ENSMUST00000021818.7
|
Cenpp
|
centromere protein P |
| chr3_+_40800013 | 1.13 |
ENSMUST00000026858.5
ENSMUST00000170825.1 |
Plk4
|
polo-like kinase 4 |
| chr1_+_58646608 | 1.13 |
ENSMUST00000081455.4
|
Gm10068
|
predicted gene 10068 |
| chr18_-_67549173 | 1.12 |
ENSMUST00000115050.1
|
Spire1
|
spire homolog 1 (Drosophila) |
| chr15_+_66670749 | 1.12 |
ENSMUST00000065916.7
|
Tg
|
thyroglobulin |
| chr9_+_65890237 | 1.07 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr11_+_78301529 | 1.07 |
ENSMUST00000045026.3
|
Spag5
|
sperm associated antigen 5 |
| chr11_-_87108656 | 1.07 |
ENSMUST00000051395.8
|
Prr11
|
proline rich 11 |
| chr17_-_53689266 | 1.06 |
ENSMUST00000024736.7
|
Sgol1
|
shugoshin-like 1 (S. pombe) |
| chr5_-_21785115 | 1.05 |
ENSMUST00000115193.1
ENSMUST00000115192.1 ENSMUST00000115195.1 ENSMUST00000030771.5 |
Dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr13_+_23763660 | 1.03 |
ENSMUST00000055770.1
|
Hist1h1a
|
histone cluster 1, H1a |
| chr18_-_43477764 | 1.03 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr1_+_139454747 | 1.01 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr4_-_99829180 | 1.01 |
ENSMUST00000146258.1
|
Itgb3bp
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr8_+_21191614 | 0.99 |
ENSMUST00000098896.4
|
Defa-rs7
|
defensin, alpha, related sequence 7 |
| chr13_+_108046411 | 0.99 |
ENSMUST00000095458.4
|
Smim15
|
small integral membrane protein 15 |
| chr2_+_131133497 | 0.97 |
ENSMUST00000110225.1
|
Gm11037
|
predicted gene 11037 |
| chr9_-_60838200 | 0.96 |
ENSMUST00000063858.7
|
Gm9869
|
predicted gene 9869 |
| chrX_-_61185558 | 0.96 |
ENSMUST00000166381.1
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr2_+_79255500 | 0.95 |
ENSMUST00000099972.4
|
Itga4
|
integrin alpha 4 |
| chr16_-_22161450 | 0.94 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr15_+_77477044 | 0.93 |
ENSMUST00000060551.2
ENSMUST00000119997.1 |
Apol10a
|
apolipoprotein L 10A |
| chr7_-_3502465 | 0.91 |
ENSMUST00000065703.7
|
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr8_+_21055047 | 0.90 |
ENSMUST00000098899.3
|
Defa23
|
defensin, alpha, 23 |
| chr5_-_67815852 | 0.89 |
ENSMUST00000141443.1
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chrX_+_94636066 | 0.89 |
ENSMUST00000096368.3
|
Gspt2
|
G1 to S phase transition 2 |
| chr2_-_164743182 | 0.88 |
ENSMUST00000103096.3
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr7_-_4752972 | 0.87 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr11_+_16951371 | 0.87 |
ENSMUST00000109635.1
ENSMUST00000061327.1 |
Fbxo48
|
F-box protein 48 |
| chr9_+_17030045 | 0.86 |
ENSMUST00000164523.2
|
Gm5611
|
predicted gene 5611 |
| chr19_-_11640828 | 0.85 |
ENSMUST00000112984.2
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr17_+_5799491 | 0.85 |
ENSMUST00000181484.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr2_+_4300462 | 0.84 |
ENSMUST00000175669.1
|
Frmd4a
|
FERM domain containing 4A |
| chr19_+_6084983 | 0.84 |
ENSMUST00000025704.2
|
Cdca5
|
cell division cycle associated 5 |
| chr6_+_8520008 | 0.83 |
ENSMUST00000162567.1
ENSMUST00000161217.1 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr10_-_13324250 | 0.81 |
ENSMUST00000105543.1
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr6_-_87672142 | 0.81 |
ENSMUST00000032130.2
ENSMUST00000065997.2 |
Aplf
|
aprataxin and PNKP like factor |
| chr9_+_64117147 | 0.81 |
ENSMUST00000034969.7
|
Lctl
|
lactase-like |
| chr9_+_53405280 | 0.81 |
ENSMUST00000005262.1
|
4930550C14Rik
|
RIKEN cDNA 4930550C14 gene |
| chr6_+_36388055 | 0.80 |
ENSMUST00000172278.1
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr5_-_149051300 | 0.78 |
ENSMUST00000110505.1
|
Hmgb1
|
high mobility group box 1 |
| chr19_+_10015016 | 0.78 |
ENSMUST00000137637.1
ENSMUST00000149967.1 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr13_+_23555023 | 0.78 |
ENSMUST00000045301.6
|
Hist1h1d
|
histone cluster 1, H1d |
| chr4_-_19922599 | 0.76 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr6_+_30723541 | 0.75 |
ENSMUST00000115127.1
|
Mest
|
mesoderm specific transcript |
| chr2_+_4400958 | 0.74 |
ENSMUST00000075767.7
|
Frmd4a
|
FERM domain containing 4A |
| chr3_+_3634145 | 0.74 |
ENSMUST00000108394.1
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr3_+_106113229 | 0.74 |
ENSMUST00000079132.5
ENSMUST00000139086.1 |
Chia
|
chitinase, acidic |
| chr10_+_3973086 | 0.74 |
ENSMUST00000117291.1
ENSMUST00000120585.1 ENSMUST00000043735.7 |
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr1_+_179961110 | 0.73 |
ENSMUST00000076687.5
ENSMUST00000097450.3 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr15_-_103252810 | 0.73 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chrX_+_75095854 | 0.73 |
ENSMUST00000033776.8
|
Dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr10_+_45577811 | 0.73 |
ENSMUST00000037044.6
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr7_+_103937382 | 0.72 |
ENSMUST00000098189.1
|
Olfr632
|
olfactory receptor 632 |
| chr12_+_77238093 | 0.72 |
ENSMUST00000177595.1
ENSMUST00000171770.2 |
Fut8
|
fucosyltransferase 8 |
| chr6_+_117863069 | 0.71 |
ENSMUST00000079405.8
ENSMUST00000172088.1 |
Zfp239
|
zinc finger protein 239 |
| chr2_+_103969528 | 0.71 |
ENSMUST00000123437.1
ENSMUST00000163256.1 |
Lmo2
|
LIM domain only 2 |
| chr13_-_62371936 | 0.70 |
ENSMUST00000107989.3
|
Gm3604
|
predicted gene 3604 |
| chr4_+_109343029 | 0.70 |
ENSMUST00000030281.5
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr8_-_123318553 | 0.70 |
ENSMUST00000118395.1
ENSMUST00000035495.8 |
Fanca
|
Fanconi anemia, complementation group A |
| chr19_-_53589067 | 0.70 |
ENSMUST00000095978.3
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chrX_+_37126777 | 0.69 |
ENSMUST00000016553.4
|
Nkap
|
NFKB activating protein |
| chr12_-_20900867 | 0.69 |
ENSMUST00000079237.5
|
Zfp125
|
zinc finger protein 125 |
| chr7_-_3915501 | 0.69 |
ENSMUST00000038176.8
ENSMUST00000090689.4 |
Lilra6
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 |
| chrX_-_160906998 | 0.68 |
ENSMUST00000069417.5
|
Gja6
|
gap junction protein, alpha 6 |
| chr6_-_125236974 | 0.68 |
ENSMUST00000112281.1
|
Cd27
|
CD27 antigen |
| chr13_-_106847267 | 0.68 |
ENSMUST00000057427.4
|
Lrrc70
|
leucine rich repeat containing 70 |
| chr3_+_19989151 | 0.68 |
ENSMUST00000173779.1
|
Cp
|
ceruloplasmin |
| chrX_+_48695004 | 0.67 |
ENSMUST00000033433.2
|
Rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr11_+_46454957 | 0.66 |
ENSMUST00000109229.1
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr4_+_141010644 | 0.66 |
ENSMUST00000071977.8
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr16_-_17125106 | 0.66 |
ENSMUST00000093336.6
|
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr6_-_125236953 | 0.66 |
ENSMUST00000112282.2
|
Cd27
|
CD27 antigen |
| chrX_+_96455359 | 0.66 |
ENSMUST00000033553.7
|
Heph
|
hephaestin |
| chr10_-_21160925 | 0.66 |
ENSMUST00000020158.6
|
Myb
|
myeloblastosis oncogene |
| chr4_+_152115934 | 0.66 |
ENSMUST00000025706.3
|
Tnfrsf25
|
tumor necrosis factor receptor superfamily, member 25 |
| chr3_-_59344256 | 0.66 |
ENSMUST00000039419.6
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr9_+_86743641 | 0.65 |
ENSMUST00000179574.1
|
Prss35
|
protease, serine, 35 |
| chr6_+_47244359 | 0.65 |
ENSMUST00000060839.6
|
Cntnap2
|
contactin associated protein-like 2 |
| chr9_+_96258697 | 0.65 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr7_-_141655319 | 0.65 |
ENSMUST00000062451.7
|
Muc6
|
mucin 6, gastric |
| chr14_+_122034660 | 0.64 |
ENSMUST00000045976.6
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chr3_-_158016419 | 0.63 |
ENSMUST00000127778.1
|
Srsf11
|
serine/arginine-rich splicing factor 11 |
| chr3_+_10366903 | 0.63 |
ENSMUST00000029049.5
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr9_+_107950952 | 0.63 |
ENSMUST00000049348.3
|
Traip
|
TRAF-interacting protein |
| chr3_-_59130610 | 0.62 |
ENSMUST00000065220.6
ENSMUST00000091112.4 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr15_+_79895017 | 0.62 |
ENSMUST00000023054.7
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr19_-_11660516 | 0.62 |
ENSMUST00000135994.1
ENSMUST00000121793.1 ENSMUST00000069681.3 |
Plac1l
|
placenta-specific 1-like |
| chr14_+_53763083 | 0.62 |
ENSMUST00000180859.1
ENSMUST00000103589.4 |
Trav14-3
|
T cell receptor alpha variable 14-3 |
| chr16_+_72663143 | 0.61 |
ENSMUST00000023600.7
|
Robo1
|
roundabout homolog 1 (Drosophila) |
| chr7_+_128129536 | 0.61 |
ENSMUST00000033053.6
|
Itgax
|
integrin alpha X |
| chr2_-_160327494 | 0.61 |
ENSMUST00000099127.2
|
Gm826
|
predicted gene 826 |
| chr17_-_81649607 | 0.60 |
ENSMUST00000163680.2
ENSMUST00000086538.3 ENSMUST00000163123.1 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr13_+_8202885 | 0.60 |
ENSMUST00000139438.1
ENSMUST00000135574.1 |
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chr19_+_33822908 | 0.60 |
ENSMUST00000042061.6
|
Gm5519
|
predicted pseudogene 5519 |
| chr8_-_126971062 | 0.60 |
ENSMUST00000045994.6
|
Rbm34
|
RNA binding motif protein 34 |
| chr13_-_104816908 | 0.60 |
ENSMUST00000022228.6
|
Cwc27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr6_+_29471437 | 0.60 |
ENSMUST00000171317.1
|
Gm9047
|
predicted gene 9047 |
| chr9_+_92542223 | 0.59 |
ENSMUST00000070522.7
ENSMUST00000160359.1 |
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr16_-_18811972 | 0.59 |
ENSMUST00000000028.7
ENSMUST00000115585.1 |
Cdc45
|
cell division cycle 45 |
| chr9_+_86743616 | 0.59 |
ENSMUST00000036426.6
|
Prss35
|
protease, serine, 35 |
| chr9_-_112187766 | 0.58 |
ENSMUST00000111872.2
ENSMUST00000164754.2 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr19_-_7802578 | 0.58 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr13_-_21440901 | 0.58 |
ENSMUST00000122872.1
ENSMUST00000151743.1 ENSMUST00000148071.1 |
Pgbd1
|
piggyBac transposable element derived 1 |
| chr1_-_181144133 | 0.57 |
ENSMUST00000027797.7
|
Nvl
|
nuclear VCP-like |
| chr4_+_152116312 | 0.57 |
ENSMUST00000035275.7
|
Tnfrsf25
|
tumor necrosis factor receptor superfamily, member 25 |
| chrX_+_6873484 | 0.56 |
ENSMUST00000145302.1
|
Dgkk
|
diacylglycerol kinase kappa |
| chrX_-_57281591 | 0.56 |
ENSMUST00000114735.2
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr3_+_124321031 | 0.56 |
ENSMUST00000058994.4
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr7_-_101864093 | 0.56 |
ENSMUST00000106981.1
|
Folr1
|
folate receptor 1 (adult) |
| chr10_-_86732409 | 0.56 |
ENSMUST00000070435.4
|
Fabp3-ps1
|
fatty acid binding protein 3, muscle and heart, pseudogene 1 |
| chr18_+_11633276 | 0.56 |
ENSMUST00000115861.2
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr16_+_11313812 | 0.55 |
ENSMUST00000023140.5
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chrX_-_134746913 | 0.55 |
ENSMUST00000096324.2
|
Gm10344
|
predicted gene 10344 |
| chr17_+_37002610 | 0.55 |
ENSMUST00000173921.1
ENSMUST00000172580.1 |
Zfp57
|
zinc finger protein 57 |
| chr7_+_70388305 | 0.55 |
ENSMUST00000080024.5
|
B130024G19Rik
|
RIKEN cDNA B130024G19 gene |
| chr5_+_90794530 | 0.55 |
ENSMUST00000031322.6
|
Cxcl15
|
chemokine (C-X-C motif) ligand 15 |
| chr6_-_128275577 | 0.54 |
ENSMUST00000130454.1
|
Tead4
|
TEA domain family member 4 |
| chr6_+_72097561 | 0.54 |
ENSMUST00000069994.4
ENSMUST00000114112.1 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr18_+_36528145 | 0.54 |
ENSMUST00000074298.6
ENSMUST00000115694.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr4_-_58499398 | 0.54 |
ENSMUST00000107570.1
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr11_+_98907801 | 0.54 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr16_+_57353093 | 0.53 |
ENSMUST00000159816.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr2_-_77946331 | 0.53 |
ENSMUST00000111821.2
ENSMUST00000111818.1 |
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr17_+_21555046 | 0.52 |
ENSMUST00000079242.3
|
Zfp52
|
zinc finger protein 52 |
| chr8_+_66386292 | 0.52 |
ENSMUST00000039540.5
ENSMUST00000110253.2 |
March1
|
membrane-associated ring finger (C3HC4) 1 |
| chr11_-_8973266 | 0.52 |
ENSMUST00000154153.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr6_-_125236996 | 0.52 |
ENSMUST00000032486.6
|
Cd27
|
CD27 antigen |
| chr3_-_102964124 | 0.52 |
ENSMUST00000058899.8
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr10_-_62602261 | 0.52 |
ENSMUST00000045866.7
|
Ddx21
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 |
| chr8_+_71887264 | 0.51 |
ENSMUST00000034259.7
|
Zfp709
|
zinc finger protein 709 |
| chr6_-_28397999 | 0.51 |
ENSMUST00000035930.4
|
Zfp800
|
zinc finger protein 800 |
| chr2_-_77946375 | 0.51 |
ENSMUST00000065889.3
|
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr16_+_88777829 | 0.50 |
ENSMUST00000009191.3
|
Gm5965
|
predicted gene 5965 |
| chr12_-_24493656 | 0.50 |
ENSMUST00000073088.2
|
Gm16372
|
predicted pseudogene 16372 |
| chrX_+_9885622 | 0.50 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr3_-_30013388 | 0.50 |
ENSMUST00000108270.3
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr2_-_114654943 | 0.50 |
ENSMUST00000028640.7
ENSMUST00000102542.3 |
Dph6
|
diphthamine biosynthesis 6 |
| chr7_+_92062392 | 0.50 |
ENSMUST00000098308.2
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr18_-_36454487 | 0.50 |
ENSMUST00000025204.5
|
Pfdn1
|
prefoldin 1 |
| chr5_+_13398688 | 0.50 |
ENSMUST00000125629.1
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_78906899 | 0.50 |
ENSMUST00000042683.6
ENSMUST00000169544.1 |
Sult6b1
|
sulfotransferase family, cytosolic, 6B, member 1 |
| chr5_-_151586924 | 0.49 |
ENSMUST00000165928.1
|
Vmn2r18
|
vomeronasal 2, receptor 18 |
| chr11_+_46235460 | 0.49 |
ENSMUST00000060185.2
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr5_-_87591582 | 0.48 |
ENSMUST00000031201.7
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr11_-_99389351 | 0.48 |
ENSMUST00000103131.4
|
Krt10
|
keratin 10 |
| chr17_-_78937031 | 0.47 |
ENSMUST00000024885.8
|
Cebpz
|
CCAAT/enhancer binding protein zeta |
| chr14_+_84443553 | 0.47 |
ENSMUST00000071370.5
|
Pcdh17
|
protocadherin 17 |
| chr10_-_10429839 | 0.47 |
ENSMUST00000045328.7
|
Adgb
|
androglobin |
| chr7_-_3845050 | 0.47 |
ENSMUST00000108615.3
ENSMUST00000119469.1 |
Pira2
|
paired-Ig-like receptor A2 |
| chr14_-_104522615 | 0.47 |
ENSMUST00000022716.2
|
Rnf219
|
ring finger protein 219 |
| chr9_+_99629496 | 0.46 |
ENSMUST00000131095.1
ENSMUST00000078367.5 ENSMUST00000112885.2 |
Dzip1l
|
DAZ interacting protein 1-like |
| chr10_-_33782115 | 0.46 |
ENSMUST00000165904.1
|
Gm4794
|
predicted gene 4794 |
| chr12_+_70154124 | 0.46 |
ENSMUST00000169156.2
ENSMUST00000182927.1 |
Abhd12b
|
abhydrolase domain containing 12B |
| chr7_+_110221697 | 0.46 |
ENSMUST00000033325.7
|
Swap70
|
SWA-70 protein |
| chr8_-_69791170 | 0.45 |
ENSMUST00000131784.1
|
Zfp866
|
zinc finger protein 866 |
| chr5_+_64092925 | 0.45 |
ENSMUST00000087324.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr5_-_100820929 | 0.45 |
ENSMUST00000117364.1
ENSMUST00000055245.6 |
Fam175a
|
family with sequence similarity 175, member A |
| chr10_-_99126321 | 0.45 |
ENSMUST00000060761.5
|
Phxr2
|
per-hexamer repeat gene 2 |
| chr3_-_146108047 | 0.45 |
ENSMUST00000160285.1
|
Wdr63
|
WD repeat domain 63 |
| chr2_+_152962485 | 0.45 |
ENSMUST00000099197.2
ENSMUST00000103155.3 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr16_+_97489994 | 0.44 |
ENSMUST00000177820.1
|
Gm9242
|
predicted pseudogene 9242 |
| chr13_-_22219820 | 0.44 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr7_+_104003259 | 0.44 |
ENSMUST00000098184.1
|
Olfr638
|
olfactory receptor 638 |
| chr2_+_150909565 | 0.43 |
ENSMUST00000028948.4
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr7_+_81523531 | 0.43 |
ENSMUST00000181903.1
|
2900076A07Rik
|
RIKEN cDNA 2900076A07 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 3.6 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.5 | 1.4 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.4 | 1.3 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.4 | 2.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 1.9 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 0.9 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 0.8 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 1.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.3 | 0.8 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.2 | 0.9 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 1.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.7 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.2 | 1.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.6 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 0.6 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 0.6 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 0.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 1.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 1.8 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.2 | 0.6 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 0.6 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.2 | 0.7 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 1.1 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.2 | 1.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 1.4 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 1.2 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 0.6 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.7 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.1 | 2.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.8 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.4 | GO:1904732 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 0.5 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.4 | GO:2000040 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 0.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.7 | GO:0046654 | 10-formyltetrahydrofolate metabolic process(GO:0009256) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 0.9 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.3 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.8 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.1 | 1.9 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.4 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 0.1 | 2.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.5 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.5 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.4 | GO:0021764 | amygdala development(GO:0021764) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 2.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.2 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 0.7 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 2.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.6 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.4 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.2 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.0 | 0.2 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 1.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 1.5 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.5 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 1.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.6 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.3 | GO:1901844 | negative regulation of cardiac muscle contraction(GO:0055118) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 1.0 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:2000642 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.0 | 1.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.8 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.6 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.6 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.6 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:1905035 | regulation of defense response to fungus(GO:1900150) regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 3.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.2 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.2 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 1.3 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0043046 | piRNA metabolic process(GO:0034587) DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.0 | 0.6 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.6 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.4 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.8 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.1 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.3 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 4.2 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.3 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.0 | 0.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 1.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 0.9 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 1.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 0.6 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 1.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.7 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 1.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 0.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 2.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 2.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.5 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.0 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 4.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0034719 | SMN complex(GO:0032797) SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 1.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.6 | 1.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.5 | 2.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 1.9 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.3 | 1.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 2.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 0.8 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 1.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.2 | 0.7 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 1.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 0.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.2 | 0.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 1.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 5.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.8 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.4 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.9 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 2.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 2.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0023023 | MHC protein complex binding(GO:0023023) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 2.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.0 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.0 | 0.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 3.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 4.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.1 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |