Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
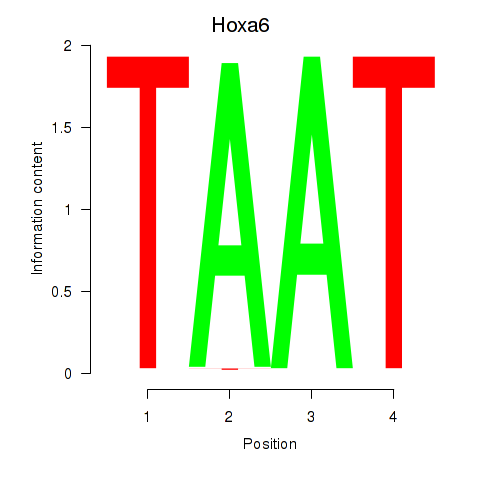
Results for Hoxa6
Z-value: 1.21
Transcription factors associated with Hoxa6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa6
|
ENSMUSG00000043219.8 | homeobox A6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa6 | mm10_v2_chr6_-_52208694_52208722 | -0.49 | 2.6e-03 | Click! |
Activity profile of Hoxa6 motif
Sorted Z-values of Hoxa6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_138079916 | 7.88 |
ENSMUST00000171804.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr2_+_70474923 | 7.16 |
ENSMUST00000100043.2
|
Sp5
|
trans-acting transcription factor 5 |
| chr16_+_42907563 | 6.89 |
ENSMUST00000151244.1
ENSMUST00000114694.2 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr14_-_118052235 | 6.45 |
ENSMUST00000022725.2
|
Dct
|
dopachrome tautomerase |
| chr1_-_150466165 | 5.96 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr8_-_5105232 | 5.85 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr2_-_148038270 | 4.86 |
ENSMUST00000132070.1
|
9030622O22Rik
|
RIKEN cDNA 9030622O22 gene |
| chr2_-_18048784 | 4.21 |
ENSMUST00000142856.1
|
Skida1
|
SKI/DACH domain containing 1 |
| chr10_+_87859255 | 4.07 |
ENSMUST00000105300.2
|
Igf1
|
insulin-like growth factor 1 |
| chr19_+_26749726 | 4.07 |
ENSMUST00000175842.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrM_+_5319 | 3.94 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr16_+_43503607 | 3.91 |
ENSMUST00000126100.1
ENSMUST00000123047.1 ENSMUST00000156981.1 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr5_+_42067960 | 3.80 |
ENSMUST00000087332.4
|
Gm16223
|
predicted gene 16223 |
| chr6_+_15185203 | 3.75 |
ENSMUST00000154448.1
|
Foxp2
|
forkhead box P2 |
| chr16_+_43247278 | 3.67 |
ENSMUST00000114691.1
ENSMUST00000079441.6 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_+_87859062 | 3.48 |
ENSMUST00000095360.4
|
Igf1
|
insulin-like growth factor 1 |
| chr14_+_29018205 | 3.33 |
ENSMUST00000055662.2
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr19_+_26605106 | 3.28 |
ENSMUST00000025862.7
ENSMUST00000176030.1 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr16_+_43235856 | 2.98 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_+_99295900 | 2.91 |
ENSMUST00000094955.1
|
Gm12689
|
predicted gene 12689 |
| chr16_+_43363855 | 2.82 |
ENSMUST00000156367.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chrM_+_7005 | 2.80 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr8_-_84773381 | 2.56 |
ENSMUST00000109764.1
|
Nfix
|
nuclear factor I/X |
| chr6_+_15185456 | 2.40 |
ENSMUST00000115472.1
ENSMUST00000115474.1 ENSMUST00000031545.7 ENSMUST00000137628.1 |
Foxp2
|
forkhead box P2 |
| chr6_-_136171722 | 2.40 |
ENSMUST00000053880.6
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr6_+_15185439 | 2.28 |
ENSMUST00000118133.1
|
Foxp2
|
forkhead box P2 |
| chr7_+_49910112 | 2.24 |
ENSMUST00000056442.5
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr13_+_63014934 | 2.21 |
ENSMUST00000091560.4
|
2010111I01Rik
|
RIKEN cDNA 2010111I01 gene |
| chr19_+_26748268 | 2.14 |
ENSMUST00000175791.1
ENSMUST00000176698.1 ENSMUST00000177252.1 ENSMUST00000176475.1 ENSMUST00000112637.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrM_+_8600 | 2.13 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr6_+_29859374 | 2.09 |
ENSMUST00000115238.3
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr9_-_61372187 | 2.07 |
ENSMUST00000098658.2
|
Gm10655
|
predicted gene 10655 |
| chr4_+_11579647 | 1.87 |
ENSMUST00000180239.1
|
Fsbp
|
fibrinogen silencer binding protein |
| chr12_-_86892540 | 1.83 |
ENSMUST00000181290.1
|
Gm26698
|
predicted gene, 26698 |
| chr13_+_40704005 | 1.82 |
ENSMUST00000069457.1
|
Gm9979
|
predicted gene 9979 |
| chr14_-_48665098 | 1.81 |
ENSMUST00000118578.1
|
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chr7_-_37773555 | 1.78 |
ENSMUST00000176534.1
|
Zfp536
|
zinc finger protein 536 |
| chr13_-_40730416 | 1.76 |
ENSMUST00000021787.5
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr4_-_42168603 | 1.76 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr14_-_48662740 | 1.73 |
ENSMUST00000122009.1
|
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chr17_-_90088343 | 1.73 |
ENSMUST00000173917.1
|
Nrxn1
|
neurexin I |
| chr14_-_12345847 | 1.70 |
ENSMUST00000022262.4
|
Fezf2
|
Fez family zinc finger 2 |
| chr5_+_115235836 | 1.70 |
ENSMUST00000081497.6
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chrM_+_10167 | 1.69 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr14_-_122451109 | 1.67 |
ENSMUST00000081580.2
|
Gm5089
|
predicted gene 5089 |
| chr16_+_43364145 | 1.66 |
ENSMUST00000148775.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_-_58052832 | 1.64 |
ENSMUST00000090940.5
|
Ermn
|
ermin, ERM-like protein |
| chr4_-_21685782 | 1.63 |
ENSMUST00000076206.4
|
Prdm13
|
PR domain containing 13 |
| chr2_-_28916412 | 1.62 |
ENSMUST00000050776.2
ENSMUST00000113849.1 |
Barhl1
|
BarH-like 1 (Drosophila) |
| chr2_-_28916668 | 1.60 |
ENSMUST00000113847.1
|
Barhl1
|
BarH-like 1 (Drosophila) |
| chr3_+_102010138 | 1.59 |
ENSMUST00000066187.4
|
Nhlh2
|
nescient helix loop helix 2 |
| chr8_+_22624019 | 1.57 |
ENSMUST00000033936.6
|
Dkk4
|
dickkopf homolog 4 (Xenopus laevis) |
| chr2_+_132847719 | 1.56 |
ENSMUST00000124836.1
ENSMUST00000154160.1 |
Crls1
|
cardiolipin synthase 1 |
| chr10_+_90576570 | 1.55 |
ENSMUST00000182786.1
ENSMUST00000182600.1 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr12_-_56613270 | 1.53 |
ENSMUST00000072631.5
|
Nkx2-9
|
NK2 homeobox 9 |
| chr4_-_14621669 | 1.53 |
ENSMUST00000143105.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr19_-_59170978 | 1.53 |
ENSMUST00000172821.2
|
Vax1
|
ventral anterior homeobox containing gene 1 |
| chr19_+_26750939 | 1.52 |
ENSMUST00000175953.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_+_43952999 | 1.50 |
ENSMUST00000177857.1
|
Rcan2
|
regulator of calcineurin 2 |
| chr5_+_90460889 | 1.49 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr10_+_90576252 | 1.48 |
ENSMUST00000182427.1
ENSMUST00000182053.1 ENSMUST00000182113.1 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_-_74659861 | 1.45 |
ENSMUST00000001867.6
|
Evx2
|
even skipped homeotic gene 2 homolog |
| chr2_+_71528657 | 1.40 |
ENSMUST00000126400.1
|
Dlx1
|
distal-less homeobox 1 |
| chr17_-_44814649 | 1.40 |
ENSMUST00000113571.3
|
Runx2
|
runt related transcription factor 2 |
| chr13_-_83729544 | 1.39 |
ENSMUST00000181705.1
|
Gm26803
|
predicted gene, 26803 |
| chr2_+_109692436 | 1.39 |
ENSMUST00000111050.3
|
Bdnf
|
brain derived neurotrophic factor |
| chr1_+_6730051 | 1.38 |
ENSMUST00000043578.6
ENSMUST00000131467.1 ENSMUST00000150761.1 ENSMUST00000151281.1 |
St18
|
suppression of tumorigenicity 18 |
| chr9_+_43744399 | 1.36 |
ENSMUST00000034510.7
|
Pvrl1
|
poliovirus receptor-related 1 |
| chr2_+_61804453 | 1.34 |
ENSMUST00000048934.8
|
Tbr1
|
T-box brain gene 1 |
| chr1_-_165934900 | 1.33 |
ENSMUST00000069609.5
ENSMUST00000111427.2 ENSMUST00000111426.4 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr3_+_118430299 | 1.32 |
ENSMUST00000180774.1
|
Gm26871
|
predicted gene, 26871 |
| chr3_-_131344892 | 1.31 |
ENSMUST00000090246.4
ENSMUST00000126569.1 |
Sgms2
|
sphingomyelin synthase 2 |
| chr3_+_115080965 | 1.30 |
ENSMUST00000051309.8
|
Olfm3
|
olfactomedin 3 |
| chr1_+_19212054 | 1.27 |
ENSMUST00000064976.4
|
Tfap2b
|
transcription factor AP-2 beta |
| chr13_-_91388079 | 1.24 |
ENSMUST00000181054.1
|
A830009L08Rik
|
RIKEN cDNA A830009L08 gene |
| chr11_+_116657106 | 1.22 |
ENSMUST00000116318.2
|
Gm11744
|
predicted gene 11744 |
| chr5_+_90518932 | 1.21 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr11_-_99521258 | 1.21 |
ENSMUST00000076948.1
|
Krt39
|
keratin 39 |
| chr12_-_31713873 | 1.20 |
ENSMUST00000057783.4
ENSMUST00000174480.2 ENSMUST00000176710.1 |
Gpr22
|
G protein-coupled receptor 22 |
| chr4_-_49408042 | 1.19 |
ENSMUST00000081541.2
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr11_+_96286623 | 1.18 |
ENSMUST00000049352.7
|
Hoxb7
|
homeobox B7 |
| chr4_-_110292719 | 1.18 |
ENSMUST00000106601.1
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chr4_-_82705735 | 1.18 |
ENSMUST00000155821.1
|
Nfib
|
nuclear factor I/B |
| chr19_+_56287911 | 1.17 |
ENSMUST00000095948.4
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr19_+_22692613 | 1.17 |
ENSMUST00000099564.2
ENSMUST00000099566.3 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr4_+_107968332 | 1.16 |
ENSMUST00000106713.3
|
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr5_-_66080971 | 1.16 |
ENSMUST00000127275.1
ENSMUST00000113724.1 |
Rbm47
|
RNA binding motif protein 47 |
| chr10_+_90576678 | 1.14 |
ENSMUST00000182284.1
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_-_24075054 | 1.14 |
ENSMUST00000068360.1
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr11_-_99521336 | 1.13 |
ENSMUST00000107445.1
|
Krt39
|
keratin 39 |
| chr4_-_110287479 | 1.13 |
ENSMUST00000106598.1
ENSMUST00000102723.4 ENSMUST00000153906.1 |
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chr10_+_90576777 | 1.12 |
ENSMUST00000183136.1
ENSMUST00000182595.1 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_-_138131356 | 1.12 |
ENSMUST00000029805.8
|
Mttp
|
microsomal triglyceride transfer protein |
| chr2_-_33942111 | 1.12 |
ENSMUST00000130988.1
ENSMUST00000127936.1 ENSMUST00000134271.1 |
Gm13403
|
predicted gene 13403 |
| chr18_-_84086379 | 1.11 |
ENSMUST00000060303.8
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr12_-_104473236 | 1.10 |
ENSMUST00000021513.4
|
Gsc
|
goosecoid homeobox |
| chr7_-_84679346 | 1.09 |
ENSMUST00000069537.2
ENSMUST00000178385.1 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr8_+_47822143 | 1.08 |
ENSMUST00000079639.2
|
Cldn24
|
claudin 24 |
| chr10_+_90576708 | 1.08 |
ENSMUST00000182430.1
ENSMUST00000182960.1 ENSMUST00000182045.1 ENSMUST00000182083.1 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_125404292 | 1.07 |
ENSMUST00000144344.1
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr2_+_116067933 | 1.05 |
ENSMUST00000156095.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr2_+_79707780 | 1.02 |
ENSMUST00000090760.2
ENSMUST00000040863.4 ENSMUST00000111780.2 |
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_-_132707304 | 1.02 |
ENSMUST00000043189.7
|
Nfasc
|
neurofascin |
| chr2_+_181767040 | 1.02 |
ENSMUST00000108756.1
|
Myt1
|
myelin transcription factor 1 |
| chr10_-_42583628 | 1.02 |
ENSMUST00000019938.4
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr1_+_187997835 | 1.02 |
ENSMUST00000110938.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr1_+_177445660 | 1.00 |
ENSMUST00000077225.6
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr16_-_88056176 | 0.99 |
ENSMUST00000072256.5
ENSMUST00000023652.8 ENSMUST00000114137.1 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr5_+_118169712 | 0.99 |
ENSMUST00000054836.6
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chrM_+_7759 | 0.99 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr3_-_37125943 | 0.98 |
ENSMUST00000029275.5
|
Il2
|
interleukin 2 |
| chr4_-_14621494 | 0.98 |
ENSMUST00000149633.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_138426735 | 0.98 |
ENSMUST00000162932.1
|
Lmo3
|
LIM domain only 3 |
| chr19_+_56287943 | 0.97 |
ENSMUST00000166049.1
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr1_+_104768510 | 0.97 |
ENSMUST00000062528.8
|
Cdh20
|
cadherin 20 |
| chr19_-_19001099 | 0.95 |
ENSMUST00000040153.8
ENSMUST00000112828.1 |
Rorb
|
RAR-related orphan receptor beta |
| chr7_-_37772868 | 0.95 |
ENSMUST00000176205.1
|
Zfp536
|
zinc finger protein 536 |
| chr7_-_49636847 | 0.95 |
ENSMUST00000032717.6
|
Dbx1
|
developing brain homeobox 1 |
| chr9_-_48964990 | 0.94 |
ENSMUST00000008734.4
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr6_+_15196949 | 0.94 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr6_+_34598530 | 0.93 |
ENSMUST00000115027.1
ENSMUST00000115026.1 |
Cald1
|
caldesmon 1 |
| chr5_-_48889531 | 0.92 |
ENSMUST00000176978.1
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr9_-_71163224 | 0.90 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chr16_+_19028232 | 0.89 |
ENSMUST00000074116.4
|
Gm10088
|
predicted gene 10088 |
| chr17_+_70561739 | 0.89 |
ENSMUST00000097288.2
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr11_+_96365752 | 0.88 |
ENSMUST00000019117.2
|
Hoxb1
|
homeobox B1 |
| chr2_-_36104060 | 0.87 |
ENSMUST00000112961.3
ENSMUST00000112966.3 |
Lhx6
|
LIM homeobox protein 6 |
| chrM_+_2743 | 0.87 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr8_-_67818218 | 0.87 |
ENSMUST00000059374.4
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_+_61372359 | 0.86 |
ENSMUST00000178113.1
ENSMUST00000159386.1 |
Tle3
|
transducin-like enhancer of split 3, homolog of Drosophila E(spl) |
| chr11_-_99374895 | 0.86 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr10_+_101681487 | 0.86 |
ENSMUST00000179929.1
ENSMUST00000127504.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr10_+_52391606 | 0.85 |
ENSMUST00000067085.4
|
Nepn
|
nephrocan |
| chr10_+_90576872 | 0.85 |
ENSMUST00000182550.1
ENSMUST00000099364.5 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_+_177444653 | 0.84 |
ENSMUST00000094276.3
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_-_63184170 | 0.84 |
ENSMUST00000112452.1
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_-_63184253 | 0.83 |
ENSMUST00000075052.3
ENSMUST00000112454.1 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr6_-_144209471 | 0.83 |
ENSMUST00000038815.7
|
Sox5
|
SRY-box containing gene 5 |
| chr2_-_64097994 | 0.83 |
ENSMUST00000131615.2
|
Fign
|
fidgetin |
| chr1_+_187997821 | 0.82 |
ENSMUST00000027906.6
|
Esrrg
|
estrogen-related receptor gamma |
| chr11_-_33203588 | 0.82 |
ENSMUST00000037746.6
|
Tlx3
|
T cell leukemia, homeobox 3 |
| chr8_-_67974567 | 0.81 |
ENSMUST00000098696.3
ENSMUST00000038959.9 ENSMUST00000093469.4 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr18_-_23041641 | 0.80 |
ENSMUST00000097651.3
|
Nol4
|
nucleolar protein 4 |
| chr2_-_35061431 | 0.80 |
ENSMUST00000028233.3
|
Hc
|
hemolytic complement |
| chr4_-_91399984 | 0.79 |
ENSMUST00000102799.3
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr2_+_74691090 | 0.79 |
ENSMUST00000061745.3
|
Hoxd10
|
homeobox D10 |
| chr9_+_44398176 | 0.78 |
ENSMUST00000165839.1
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr14_-_48667508 | 0.78 |
ENSMUST00000144465.1
ENSMUST00000133479.1 ENSMUST00000119070.1 ENSMUST00000152018.1 |
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chr16_-_92400067 | 0.78 |
ENSMUST00000023672.8
|
Rcan1
|
regulator of calcineurin 1 |
| chr6_+_104493220 | 0.78 |
ENSMUST00000162872.1
|
Cntn6
|
contactin 6 |
| chr2_+_74721978 | 0.77 |
ENSMUST00000047904.3
|
Hoxd3
|
homeobox D3 |
| chr1_+_81077204 | 0.77 |
ENSMUST00000123720.1
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr6_-_144209448 | 0.77 |
ENSMUST00000077160.5
|
Sox5
|
SRY-box containing gene 5 |
| chr2_+_20737306 | 0.77 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr13_-_18382041 | 0.76 |
ENSMUST00000139064.2
ENSMUST00000175703.2 |
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr6_+_104492790 | 0.76 |
ENSMUST00000161446.1
ENSMUST00000161070.1 ENSMUST00000089215.5 |
Cntn6
|
contactin 6 |
| chr11_+_100320596 | 0.74 |
ENSMUST00000152521.1
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr1_+_6730135 | 0.74 |
ENSMUST00000155921.1
|
St18
|
suppression of tumorigenicity 18 |
| chr11_-_100088226 | 0.74 |
ENSMUST00000107419.1
|
Krt32
|
keratin 32 |
| chr9_+_43829963 | 0.74 |
ENSMUST00000180221.1
|
Gm3898
|
predicted gene 3898 |
| chr6_-_144209558 | 0.72 |
ENSMUST00000111749.1
ENSMUST00000170367.2 |
Sox5
|
SRY-box containing gene 5 |
| chr11_-_42000532 | 0.71 |
ENSMUST00000070735.3
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr1_+_6734827 | 0.71 |
ENSMUST00000139838.1
|
St18
|
suppression of tumorigenicity 18 |
| chr19_-_45235811 | 0.70 |
ENSMUST00000099401.4
|
Lbx1
|
ladybird homeobox homolog 1 (Drosophila) |
| chr4_-_120815703 | 0.69 |
ENSMUST00000120779.1
|
Nfyc
|
nuclear transcription factor-Y gamma |
| chr11_+_78826575 | 0.69 |
ENSMUST00000147875.2
ENSMUST00000141321.1 |
Lyrm9
|
LYR motif containing 9 |
| chr5_-_67099045 | 0.69 |
ENSMUST00000174251.1
|
Phox2b
|
paired-like homeobox 2b |
| chr6_+_34598500 | 0.69 |
ENSMUST00000079391.3
ENSMUST00000142512.1 |
Cald1
|
caldesmon 1 |
| chr14_-_124677089 | 0.69 |
ENSMUST00000095529.3
|
Fgf14
|
fibroblast growth factor 14 |
| chr9_-_37669170 | 0.66 |
ENSMUST00000011262.2
|
Panx3
|
pannexin 3 |
| chr11_-_42000834 | 0.65 |
ENSMUST00000070725.4
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr11_+_96292453 | 0.65 |
ENSMUST00000173432.1
|
Hoxb6
|
homeobox B6 |
| chr8_-_109251698 | 0.64 |
ENSMUST00000079189.3
|
4922502B01Rik
|
RIKEN cDNA 4922502B01 gene |
| chr9_-_87731248 | 0.64 |
ENSMUST00000034991.7
|
Tbx18
|
T-box18 |
| chr6_+_29859662 | 0.62 |
ENSMUST00000128927.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr1_-_12991109 | 0.62 |
ENSMUST00000115403.2
ENSMUST00000115402.1 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr10_-_92164666 | 0.62 |
ENSMUST00000183123.1
ENSMUST00000182033.1 |
Rmst
|
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
| chr8_-_67818284 | 0.61 |
ENSMUST00000120071.1
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_+_34709442 | 0.61 |
ENSMUST00000115021.1
|
Cald1
|
caldesmon 1 |
| chr11_+_94044331 | 0.60 |
ENSMUST00000024979.8
|
Spag9
|
sperm associated antigen 9 |
| chr13_+_80883403 | 0.59 |
ENSMUST00000099356.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr4_-_97584605 | 0.59 |
ENSMUST00000107067.1
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr6_+_96113146 | 0.59 |
ENSMUST00000122120.1
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr9_+_46998931 | 0.58 |
ENSMUST00000178065.1
|
Gm4791
|
predicted gene 4791 |
| chr13_+_42681513 | 0.58 |
ENSMUST00000149235.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr2_+_101624696 | 0.57 |
ENSMUST00000044031.3
|
Rag2
|
recombination activating gene 2 |
| chr3_+_125404072 | 0.57 |
ENSMUST00000173932.1
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chrX_-_43274786 | 0.56 |
ENSMUST00000016294.7
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr5_-_106574706 | 0.56 |
ENSMUST00000131029.1
ENSMUST00000124394.2 |
RP24-421H18.1
|
RP24-421H18.1 |
| chr9_-_105521147 | 0.55 |
ENSMUST00000176770.1
ENSMUST00000085133.6 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr9_+_32116040 | 0.54 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr6_-_138421379 | 0.53 |
ENSMUST00000163065.1
|
Lmo3
|
LIM domain only 3 |
| chr6_+_80018877 | 0.53 |
ENSMUST00000147663.1
ENSMUST00000128718.1 ENSMUST00000126005.1 ENSMUST00000133918.1 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr2_-_76889753 | 0.53 |
ENSMUST00000148747.1
|
Ttn
|
titin |
| chr9_+_118478182 | 0.53 |
ENSMUST00000111763.1
|
Eomes
|
eomesodermin homolog (Xenopus laevis) |
| chr13_-_53473074 | 0.52 |
ENSMUST00000021922.8
|
Msx2
|
msh homeobox 2 |
| chr3_+_93393696 | 0.52 |
ENSMUST00000045912.2
|
Rptn
|
repetin |
| chr6_-_52165413 | 0.52 |
ENSMUST00000014848.8
|
Hoxa2
|
homeobox A2 |
| chr13_+_55399648 | 0.52 |
ENSMUST00000057167.7
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr3_-_26133734 | 0.50 |
ENSMUST00000108308.3
ENSMUST00000075054.4 |
Nlgn1
|
neuroligin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 1.3 | 7.5 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.2 | 11.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.7 | 6.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.7 | 4.3 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.6 | 7.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.6 | 2.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.6 | 1.8 | GO:0021622 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.6 | 9.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.6 | 2.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.5 | 1.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.5 | 2.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.5 | 1.4 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.5 | 1.4 | GO:0061193 | taste bud development(GO:0061193) |
| 0.4 | 21.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.4 | 1.7 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.4 | 2.6 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.4 | 1.3 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.4 | 3.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 3.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 0.9 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.3 | 1.2 | GO:2000795 | lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 2.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 1.4 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.3 | 3.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 7.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.3 | 1.9 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.3 | 0.8 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.3 | 0.8 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 0.2 | 6.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 2.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.2 | 1.5 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 1.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 1.0 | GO:0071205 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 | 0.6 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 1.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 2.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.5 | GO:0060364 | embryonic nail plate morphogenesis(GO:0035880) regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.2 | 0.5 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 2.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.0 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.2 | 1.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.2 | 0.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 1.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 0.8 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 0.5 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 2.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0070172 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 1.0 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.7 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.5 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 1.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.4 | GO:1900108 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.5 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 4.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 3.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0061324 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) |
| 0.1 | 1.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 2.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.4 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 1.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) |
| 0.0 | 2.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.6 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 1.1 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.0 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 1.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 1.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 2.1 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 1.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 2.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 1.6 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.5 | GO:0042567 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 0.6 | 11.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 6.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 1.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 1.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 6.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 2.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 2.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.3 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 1.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 7.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 6.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 2.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 1.7 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.5 | 1.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.4 | 3.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 1.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.4 | 2.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 6.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 8.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 0.9 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.3 | 2.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.3 | 1.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 6.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 7.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 0.8 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 0.9 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.2 | 3.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 11.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 2.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.6 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.1 | 0.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 1.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 24.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 2.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 4.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.6 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 1.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 2.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.2 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 2.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 2.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 1.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 7.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.6 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.7 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 11.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 3.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |