Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
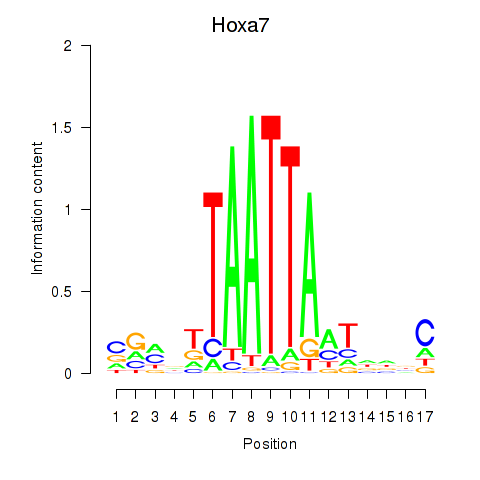
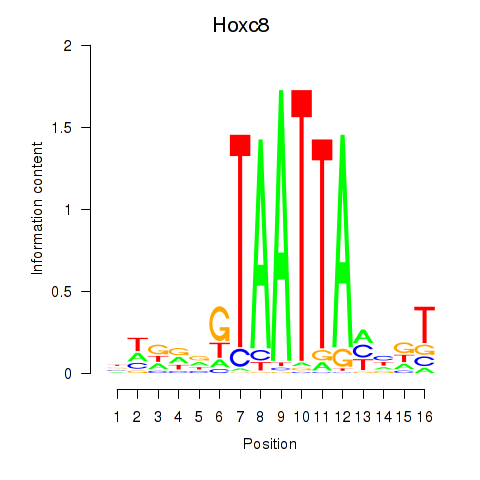
Results for Hoxa7_Hoxc8
Z-value: 0.38
Transcription factors associated with Hoxa7_Hoxc8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa7
|
ENSMUSG00000038236.6 | homeobox A7 |
|
Hoxc8
|
ENSMUSG00000001657.6 | homeobox C8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc8 | mm10_v2_chr15_+_102990576_102990622 | -0.21 | 2.2e-01 | Click! |
| Hoxa7 | mm10_v2_chr6_-_52218686_52218716 | -0.20 | 2.4e-01 | Click! |
Activity profile of Hoxa7_Hoxc8 motif
Sorted Z-values of Hoxa7_Hoxc8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_80000292 | 1.09 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr6_+_30541582 | 1.04 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr3_-_75270073 | 0.62 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr2_+_69897255 | 0.43 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr9_-_90255927 | 0.39 |
ENSMUST00000144646.1
|
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr14_-_100149764 | 0.32 |
ENSMUST00000097079.4
|
Klf12
|
Kruppel-like factor 12 |
| chr15_+_25773985 | 0.32 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr4_+_108719649 | 0.32 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr12_-_55014329 | 0.30 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr3_+_5218516 | 0.30 |
ENSMUST00000175866.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr3_-_81924463 | 0.29 |
ENSMUST00000069880.2
|
Gm9989
|
predicted gene 9989 |
| chr3_+_103832562 | 0.26 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr15_+_98571004 | 0.25 |
ENSMUST00000023728.6
|
4930415O20Rik
|
RIKEN cDNA 4930415O20 gene |
| chr17_+_23853519 | 0.24 |
ENSMUST00000061725.7
|
Prss32
|
protease, serine, 32 |
| chr4_+_34893772 | 0.24 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr12_-_40248073 | 0.23 |
ENSMUST00000169926.1
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr2_+_69219971 | 0.23 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr5_-_62766153 | 0.23 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr8_+_94152607 | 0.23 |
ENSMUST00000034211.8
|
Mt3
|
metallothionein 3 |
| chr4_-_82850721 | 0.22 |
ENSMUST00000139401.1
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr2_-_121235689 | 0.22 |
ENSMUST00000142400.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr15_-_75905349 | 0.21 |
ENSMUST00000127550.1
|
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr5_-_62765618 | 0.21 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr13_-_102905740 | 0.20 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_-_45470201 | 0.19 |
ENSMUST00000079390.6
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr14_+_27000362 | 0.19 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr13_-_102906046 | 0.19 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_50678642 | 0.19 |
ENSMUST00000072685.6
ENSMUST00000164039.2 |
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr11_-_99374895 | 0.18 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr13_+_23575753 | 0.18 |
ENSMUST00000105105.1
|
Hist1h3d
|
histone cluster 1, H3d |
| chr1_+_72284367 | 0.17 |
ENSMUST00000027380.5
ENSMUST00000141783.1 |
Tmem169
|
transmembrane protein 169 |
| chr1_-_134955847 | 0.16 |
ENSMUST00000168381.1
|
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr10_+_37139558 | 0.16 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr15_-_100425050 | 0.15 |
ENSMUST00000123461.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr3_-_59220150 | 0.15 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr1_-_126738167 | 0.15 |
ENSMUST00000160693.1
|
Nckap5
|
NCK-associated protein 5 |
| chr7_+_45621805 | 0.14 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr14_-_36919314 | 0.14 |
ENSMUST00000182797.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr15_-_37458523 | 0.14 |
ENSMUST00000116445.2
|
Ncald
|
neurocalcin delta |
| chr4_+_145585166 | 0.14 |
ENSMUST00000105739.1
ENSMUST00000119718.1 |
Gm13212
|
predicted gene 13212 |
| chr15_+_81744848 | 0.13 |
ENSMUST00000109554.1
|
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chrX_+_159303266 | 0.13 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr8_+_45885479 | 0.13 |
ENSMUST00000034053.5
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr4_+_102589687 | 0.13 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chrX_+_114474569 | 0.13 |
ENSMUST00000133447.1
|
Klhl4
|
kelch-like 4 |
| chr15_-_8710734 | 0.12 |
ENSMUST00000005493.7
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr9_-_55919605 | 0.12 |
ENSMUST00000037408.8
|
Scaper
|
S phase cyclin A-associated protein in the ER |
| chr6_-_118479237 | 0.12 |
ENSMUST00000161170.1
|
Zfp9
|
zinc finger protein 9 |
| chr3_+_5218589 | 0.12 |
ENSMUST00000177488.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr11_+_31832660 | 0.12 |
ENSMUST00000132857.1
|
Gm12107
|
predicted gene 12107 |
| chr3_+_121291725 | 0.12 |
ENSMUST00000039442.7
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr9_+_119063429 | 0.11 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr2_-_45117349 | 0.11 |
ENSMUST00000176438.2
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_34039437 | 0.11 |
ENSMUST00000131134.1
ENSMUST00000087497.4 ENSMUST00000114255.1 ENSMUST00000114252.1 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr10_-_20724696 | 0.11 |
ENSMUST00000170265.1
|
Pde7b
|
phosphodiesterase 7B |
| chr6_-_130193112 | 0.11 |
ENSMUST00000112032.1
ENSMUST00000071554.2 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr3_+_59952185 | 0.10 |
ENSMUST00000094227.3
|
Gm9696
|
predicted gene 9696 |
| chr9_+_72806874 | 0.09 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr9_+_113812547 | 0.09 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr13_-_55021196 | 0.09 |
ENSMUST00000153665.1
|
Hk3
|
hexokinase 3 |
| chr3_+_96645579 | 0.09 |
ENSMUST00000119365.1
ENSMUST00000029744.5 |
Itga10
|
integrin, alpha 10 |
| chr2_+_110721340 | 0.09 |
ENSMUST00000111016.2
|
Muc15
|
mucin 15 |
| chr19_-_53944621 | 0.09 |
ENSMUST00000135402.2
|
Bbip1
|
BBSome interacting protein 1 |
| chr1_-_132367879 | 0.09 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr10_-_76110956 | 0.08 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr11_+_29373618 | 0.08 |
ENSMUST00000040182.6
ENSMUST00000109477.1 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr3_-_86548268 | 0.08 |
ENSMUST00000077524.3
|
Mab21l2
|
mab-21-like 2 (C. elegans) |
| chr2_-_116067391 | 0.08 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr10_-_117148474 | 0.08 |
ENSMUST00000020381.3
|
Frs2
|
fibroblast growth factor receptor substrate 2 |
| chr2_-_164171113 | 0.08 |
ENSMUST00000045196.3
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr3_+_55782500 | 0.08 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr19_-_32196393 | 0.08 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr18_+_53551594 | 0.07 |
ENSMUST00000115398.1
|
Prdm6
|
PR domain containing 6 |
| chr2_+_36230426 | 0.07 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr16_-_75909272 | 0.07 |
ENSMUST00000114239.2
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr6_+_37870786 | 0.07 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr18_+_4920509 | 0.07 |
ENSMUST00000126977.1
|
Svil
|
supervillin |
| chr7_+_30493622 | 0.07 |
ENSMUST00000058280.6
ENSMUST00000133318.1 ENSMUST00000142575.1 ENSMUST00000131040.1 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr7_+_126950687 | 0.07 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr19_-_55241236 | 0.06 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr18_+_37518341 | 0.06 |
ENSMUST00000097609.1
|
Pcdhb22
|
protocadherin beta 22 |
| chr14_+_54431597 | 0.06 |
ENSMUST00000089688.4
|
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr16_-_50330987 | 0.06 |
ENSMUST00000114488.1
|
Bbx
|
bobby sox homolog (Drosophila) |
| chr17_-_45659312 | 0.06 |
ENSMUST00000120717.1
|
Capn11
|
calpain 11 |
| chr10_-_86011833 | 0.06 |
ENSMUST00000105304.1
ENSMUST00000061699.5 |
Bpifc
|
BPI fold containing family C |
| chr10_+_76147451 | 0.06 |
ENSMUST00000020450.3
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr3_+_5214763 | 0.06 |
ENSMUST00000177924.1
|
Gm10748
|
predicted gene 10748 |
| chr9_-_79977782 | 0.06 |
ENSMUST00000093811.4
|
Filip1
|
filamin A interacting protein 1 |
| chr6_-_30693676 | 0.06 |
ENSMUST00000169422.1
ENSMUST00000115131.1 ENSMUST00000115130.2 ENSMUST00000031810.8 |
Cep41
|
centrosomal protein 41 |
| chr4_-_115133977 | 0.06 |
ENSMUST00000051400.7
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr3_-_17230976 | 0.05 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr2_-_79456750 | 0.05 |
ENSMUST00000041099.4
|
Neurod1
|
neurogenic differentiation 1 |
| chrM_+_9870 | 0.05 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr8_-_24725762 | 0.05 |
ENSMUST00000171438.1
ENSMUST00000171611.1 ENSMUST00000033958.7 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr18_-_56975333 | 0.05 |
ENSMUST00000139243.2
ENSMUST00000025488.8 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr12_+_98746935 | 0.05 |
ENSMUST00000110105.2
ENSMUST00000110104.2 ENSMUST00000057000.9 |
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr7_-_46667303 | 0.05 |
ENSMUST00000168335.1
|
Tph1
|
tryptophan hydroxylase 1 |
| chr5_+_66968559 | 0.04 |
ENSMUST00000127184.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr5_+_138187485 | 0.04 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr2_-_177267036 | 0.04 |
ENSMUST00000108963.1
|
Gm14409
|
predicted gene 14409 |
| chr16_-_45724600 | 0.04 |
ENSMUST00000096057.4
|
Tagln3
|
transgelin 3 |
| chr2_+_23069210 | 0.04 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr1_-_24612700 | 0.04 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr12_+_71170589 | 0.04 |
ENSMUST00000129376.1
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr10_+_41303968 | 0.04 |
ENSMUST00000173517.2
|
Ak9
|
adenylate kinase 9 |
| chr18_+_23753708 | 0.04 |
ENSMUST00000115830.1
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_-_172027269 | 0.03 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr2_+_83724397 | 0.03 |
ENSMUST00000028499.4
ENSMUST00000141725.1 ENSMUST00000111740.2 |
Itgav
|
integrin alpha V |
| chr13_-_81710937 | 0.03 |
ENSMUST00000161920.1
ENSMUST00000048993.5 |
Polr3g
|
polymerase (RNA) III (DNA directed) polypeptide G |
| chr3_+_5218546 | 0.03 |
ENSMUST00000026284.6
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr19_+_5474681 | 0.03 |
ENSMUST00000165485.1
ENSMUST00000166253.1 ENSMUST00000167371.1 ENSMUST00000167855.1 ENSMUST00000070118.7 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr15_-_8710409 | 0.03 |
ENSMUST00000157065.1
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_+_104202609 | 0.03 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr16_+_45224315 | 0.03 |
ENSMUST00000102802.3
ENSMUST00000063654.4 |
Btla
|
B and T lymphocyte associated |
| chr1_-_150164943 | 0.03 |
ENSMUST00000181308.1
|
Gm26687
|
predicted gene, 26687 |
| chr1_-_134955908 | 0.02 |
ENSMUST00000045665.6
ENSMUST00000086444.4 ENSMUST00000112163.1 |
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chrX_-_101086020 | 0.02 |
ENSMUST00000113710.1
|
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr3_+_55461758 | 0.02 |
ENSMUST00000070418.4
|
Dclk1
|
doublecortin-like kinase 1 |
| chr6_-_137169710 | 0.02 |
ENSMUST00000117919.1
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr19_-_41933276 | 0.02 |
ENSMUST00000075280.4
ENSMUST00000112123.2 |
Exosc1
|
exosome component 1 |
| chr2_+_127854628 | 0.02 |
ENSMUST00000028859.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chrX_+_166170449 | 0.01 |
ENSMUST00000130880.2
ENSMUST00000056410.4 ENSMUST00000096252.3 ENSMUST00000087169.4 |
Gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr11_-_73326472 | 0.01 |
ENSMUST00000155630.2
|
Aspa
|
aspartoacylase |
| chr11_-_65269941 | 0.01 |
ENSMUST00000102635.3
|
Myocd
|
myocardin |
| chr9_+_38877126 | 0.01 |
ENSMUST00000078289.2
|
Olfr926
|
olfactory receptor 926 |
| chr15_-_67113909 | 0.01 |
ENSMUST00000092640.5
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr4_-_136602641 | 0.01 |
ENSMUST00000105847.1
ENSMUST00000116273.2 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr17_+_88440711 | 0.01 |
ENSMUST00000112238.2
ENSMUST00000155640.1 |
Foxn2
|
forkhead box N2 |
| chr8_+_23411490 | 0.01 |
ENSMUST00000033952.7
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr1_+_82316452 | 0.00 |
ENSMUST00000027322.7
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr18_+_55057557 | 0.00 |
ENSMUST00000181765.1
|
Gm4221
|
predicted gene 4221 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa7_Hoxc8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 1.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.0 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) integrin alphav-beta6 complex(GO:0034685) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |