Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxb2_Dlx2
Z-value: 0.65
Transcription factors associated with Hoxb2_Dlx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
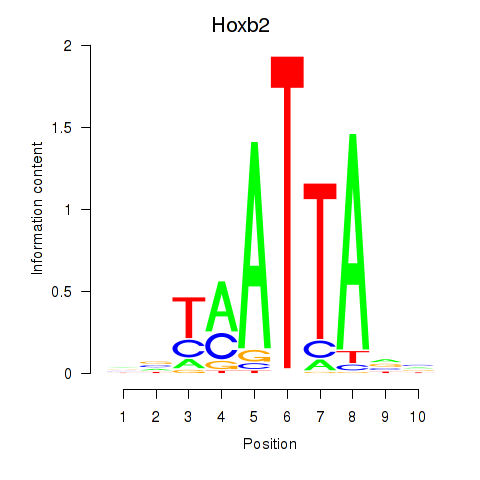
Hoxb2
|
ENSMUSG00000075588.5 | homeobox B2 |
|
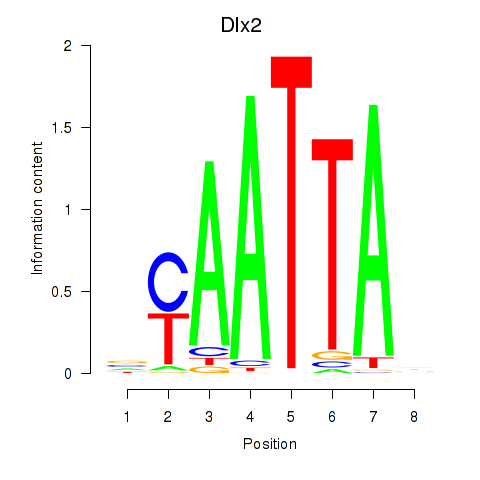
Dlx2
|
ENSMUSG00000023391.7 | distal-less homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx2 | mm10_v2_chr2_-_71546745_71546758 | -0.61 | 6.6e-05 | Click! |
| Hoxb2 | mm10_v2_chr11_+_96351632_96351632 | -0.60 | 9.6e-05 | Click! |
Activity profile of Hoxb2_Dlx2 motif
Sorted Z-values of Hoxb2_Dlx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_60081861 | 7.01 |
ENSMUST00000029326.5
|
Sucnr1
|
succinate receptor 1 |
| chr13_+_4434306 | 5.07 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr19_-_7966000 | 4.53 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr6_-_138079916 | 4.34 |
ENSMUST00000171804.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr3_+_62419668 | 4.27 |
ENSMUST00000161057.1
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr10_+_127898515 | 3.52 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr10_-_92375367 | 3.43 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr2_+_68104671 | 3.37 |
ENSMUST00000042456.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr15_-_60921270 | 3.12 |
ENSMUST00000096418.3
|
A1bg
|
alpha-1-B glycoprotein |
| chrX_-_143933089 | 2.94 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr14_+_33941021 | 2.91 |
ENSMUST00000100720.1
|
Gdf2
|
growth differentiation factor 2 |
| chr10_-_24101951 | 2.90 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr16_+_42907563 | 2.85 |
ENSMUST00000151244.1
ENSMUST00000114694.2 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_72284248 | 2.54 |
ENSMUST00000097698.4
ENSMUST00000027381.6 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr9_-_71163224 | 2.42 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chrM_+_10167 | 2.36 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_-_140183283 | 2.33 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr1_-_139608282 | 2.32 |
ENSMUST00000170441.2
|
Cfhr3
|
complement factor H-related 3 |
| chr1_-_140183404 | 2.29 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr5_-_87569023 | 2.29 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr10_-_107486077 | 2.25 |
ENSMUST00000000445.1
|
Myf5
|
myogenic factor 5 |
| chr14_-_118052235 | 2.22 |
ENSMUST00000022725.2
|
Dct
|
dopachrome tautomerase |
| chr2_+_116067213 | 2.15 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr5_+_90561102 | 2.15 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr2_+_70474923 | 2.09 |
ENSMUST00000100043.2
|
Sp5
|
trans-acting transcription factor 5 |
| chr7_+_103550368 | 2.02 |
ENSMUST00000106888.1
|
Olfr613
|
olfactory receptor 613 |
| chr16_+_22918378 | 2.00 |
ENSMUST00000170805.1
|
Fetub
|
fetuin beta |
| chrM_+_9870 | 1.94 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrM_+_7005 | 1.89 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr3_+_138374121 | 1.88 |
ENSMUST00000171054.1
|
Adh6-ps1
|
alcohol dehydrogenase 6 (class V), pseudogene 1 |
| chr5_+_139543889 | 1.80 |
ENSMUST00000174792.1
ENSMUST00000031523.8 |
Uncx
|
UNC homeobox |
| chr7_-_48848023 | 1.80 |
ENSMUST00000032658.6
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr3_+_138313279 | 1.77 |
ENSMUST00000013455.6
ENSMUST00000106247.1 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr7_-_100656953 | 1.70 |
ENSMUST00000107046.1
ENSMUST00000107045.1 ENSMUST00000139708.1 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr1_-_24612700 | 1.67 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr3_+_122419772 | 1.59 |
ENSMUST00000029766.4
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr5_-_87482258 | 1.56 |
ENSMUST00000079811.6
ENSMUST00000144144.1 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr10_+_39612934 | 1.56 |
ENSMUST00000019987.6
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr2_+_69897220 | 1.55 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr11_-_99521258 | 1.54 |
ENSMUST00000076948.1
|
Krt39
|
keratin 39 |
| chr14_-_68533689 | 1.53 |
ENSMUST00000022640.7
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr5_+_42067960 | 1.49 |
ENSMUST00000087332.4
|
Gm16223
|
predicted gene 16223 |
| chr8_-_5105232 | 1.49 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr4_+_108719649 | 1.47 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr17_-_34882094 | 1.47 |
ENSMUST00000025230.8
|
C2
|
complement component 2 (within H-2S) |
| chr18_-_75697639 | 1.46 |
ENSMUST00000165559.1
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr1_+_88200601 | 1.45 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr11_-_113710017 | 1.44 |
ENSMUST00000018871.1
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr15_-_101892916 | 1.42 |
ENSMUST00000100179.1
|
Krt76
|
keratin 76 |
| chr3_+_133338936 | 1.40 |
ENSMUST00000150386.1
ENSMUST00000125858.1 |
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr6_-_115592571 | 1.39 |
ENSMUST00000112957.1
|
2510049J12Rik
|
RIKEN cDNA 2510049J12 gene |
| chr11_-_99521336 | 1.38 |
ENSMUST00000107445.1
|
Krt39
|
keratin 39 |
| chr17_-_83846769 | 1.37 |
ENSMUST00000000687.7
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr11_-_43901187 | 1.36 |
ENSMUST00000067258.2
ENSMUST00000139906.1 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr5_-_62766153 | 1.34 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr15_-_37458523 | 1.33 |
ENSMUST00000116445.2
|
Ncald
|
neurocalcin delta |
| chr6_+_124304646 | 1.30 |
ENSMUST00000112541.2
ENSMUST00000032234.2 |
Cd163
|
CD163 antigen |
| chr11_-_99374895 | 1.27 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr19_-_39649046 | 1.26 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr9_-_71168657 | 1.26 |
ENSMUST00000113570.1
|
Aqp9
|
aquaporin 9 |
| chrX_-_60893430 | 1.26 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr8_+_45658666 | 1.26 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr16_+_43235856 | 1.25 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_+_43829963 | 1.24 |
ENSMUST00000180221.1
|
Gm3898
|
predicted gene 3898 |
| chrX_-_143933204 | 1.24 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr4_-_104876383 | 1.23 |
ENSMUST00000064873.8
ENSMUST00000106808.3 ENSMUST00000048947.8 |
C8a
|
complement component 8, alpha polypeptide |
| chr5_-_86373413 | 1.21 |
ENSMUST00000031175.5
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr2_-_5676046 | 1.19 |
ENSMUST00000114987.3
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr2_-_166155624 | 1.19 |
ENSMUST00000109249.2
|
Sulf2
|
sulfatase 2 |
| chr2_+_109917639 | 1.19 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr14_+_52824340 | 1.17 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr9_+_32116040 | 1.15 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr18_-_38866702 | 1.14 |
ENSMUST00000115582.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr12_-_57546121 | 1.13 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr7_-_34654342 | 1.13 |
ENSMUST00000108069.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chrM_+_9452 | 1.13 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr6_+_8948608 | 1.11 |
ENSMUST00000160300.1
|
Nxph1
|
neurexophilin 1 |
| chr13_+_89540636 | 1.09 |
ENSMUST00000022108.7
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_+_74284930 | 1.08 |
ENSMUST00000113805.1
ENSMUST00000027370.6 ENSMUST00000087226.4 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr12_-_56613270 | 1.07 |
ENSMUST00000072631.5
|
Nkx2-9
|
NK2 homeobox 9 |
| chr2_-_166155272 | 1.07 |
ENSMUST00000088086.3
|
Sulf2
|
sulfatase 2 |
| chr3_-_157925056 | 1.06 |
ENSMUST00000118539.1
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr2_-_37359274 | 1.05 |
ENSMUST00000009174.8
|
Pdcl
|
phosducin-like |
| chr15_-_67113909 | 1.03 |
ENSMUST00000092640.5
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr12_-_48559971 | 1.03 |
ENSMUST00000169406.1
|
Gm1818
|
predicted gene 1818 |
| chr7_+_5015466 | 1.03 |
ENSMUST00000086349.3
|
Zfp524
|
zinc finger protein 524 |
| chr9_-_15301555 | 1.03 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_+_99873389 | 1.03 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr13_+_23807027 | 1.02 |
ENSMUST00000006786.4
ENSMUST00000099697.2 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr8_-_84846860 | 1.01 |
ENSMUST00000003912.6
|
Calr
|
calreticulin |
| chr19_+_26749726 | 0.98 |
ENSMUST00000175842.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_+_87925624 | 0.97 |
ENSMUST00000113271.2
|
Csn3
|
casein kappa |
| chr2_+_23069210 | 0.96 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr18_-_3281036 | 0.96 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr19_-_42202150 | 0.95 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr11_-_119547744 | 0.94 |
ENSMUST00000026670.4
|
Nptx1
|
neuronal pentraxin 1 |
| chr3_-_19264959 | 0.94 |
ENSMUST00000121951.1
|
Pde7a
|
phosphodiesterase 7A |
| chr9_+_121950988 | 0.94 |
ENSMUST00000043011.7
|
Fam198a
|
family with sequence similarity 198, member A |
| chr4_+_97777780 | 0.94 |
ENSMUST00000107062.2
ENSMUST00000052018.5 ENSMUST00000107057.1 |
Nfia
|
nuclear factor I/A |
| chr18_+_56432116 | 0.91 |
ENSMUST00000070166.5
|
Gramd3
|
GRAM domain containing 3 |
| chr2_-_77703252 | 0.91 |
ENSMUST00000171063.1
|
Zfp385b
|
zinc finger protein 385B |
| chr1_+_132298606 | 0.91 |
ENSMUST00000046071.4
|
Klhdc8a
|
kelch domain containing 8A |
| chr16_+_37580137 | 0.90 |
ENSMUST00000160847.1
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr17_-_90088343 | 0.90 |
ENSMUST00000173917.1
|
Nrxn1
|
neurexin I |
| chr6_-_125380793 | 0.90 |
ENSMUST00000042647.6
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chrX_-_100594860 | 0.89 |
ENSMUST00000053373.1
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr4_+_136143497 | 0.88 |
ENSMUST00000008016.2
|
Id3
|
inhibitor of DNA binding 3 |
| chr17_+_45734506 | 0.87 |
ENSMUST00000180558.1
|
F630040K05Rik
|
RIKEN cDNA F630040K05 gene |
| chrX_+_57212110 | 0.84 |
ENSMUST00000033466.1
|
Cd40lg
|
CD40 ligand |
| chr13_-_53473074 | 0.83 |
ENSMUST00000021922.8
|
Msx2
|
msh homeobox 2 |
| chr2_+_132847719 | 0.83 |
ENSMUST00000124836.1
ENSMUST00000154160.1 |
Crls1
|
cardiolipin synthase 1 |
| chr6_+_37870786 | 0.83 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr10_+_53337686 | 0.83 |
ENSMUST00000046221.6
ENSMUST00000163319.1 |
Pln
|
phospholamban |
| chr12_-_104473236 | 0.82 |
ENSMUST00000021513.4
|
Gsc
|
goosecoid homeobox |
| chr3_+_94398517 | 0.81 |
ENSMUST00000050975.3
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr14_+_55559993 | 0.81 |
ENSMUST00000117236.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_-_14621669 | 0.80 |
ENSMUST00000143105.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_-_62765618 | 0.80 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr14_+_55560010 | 0.79 |
ENSMUST00000147981.1
ENSMUST00000133256.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr6_-_136171722 | 0.78 |
ENSMUST00000053880.6
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr2_-_71750083 | 0.78 |
ENSMUST00000180494.1
|
Gm17250
|
predicted gene, 17250 |
| chr6_-_98342728 | 0.78 |
ENSMUST00000164491.1
|
Gm765
|
predicted gene 765 |
| chr2_+_20737306 | 0.78 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr18_+_9707639 | 0.78 |
ENSMUST00000040069.8
|
Colec12
|
collectin sub-family member 12 |
| chr2_+_69897255 | 0.76 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr9_-_16378231 | 0.75 |
ENSMUST00000082170.5
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila) |
| chr11_+_99879187 | 0.74 |
ENSMUST00000078442.3
|
Gm11567
|
predicted gene 11567 |
| chr1_-_163725123 | 0.74 |
ENSMUST00000159679.1
|
Mettl11b
|
methyltransferase like 11B |
| chr11_+_73350839 | 0.73 |
ENSMUST00000120137.1
|
Olfr20
|
olfactory receptor 20 |
| chr6_-_130231638 | 0.73 |
ENSMUST00000088011.4
ENSMUST00000112013.1 ENSMUST00000049304.7 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr17_+_36958571 | 0.73 |
ENSMUST00000040177.6
|
Znrd1as
|
Znrd1 antisense |
| chr8_+_45658731 | 0.73 |
ENSMUST00000143820.1
ENSMUST00000132139.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_+_31224371 | 0.73 |
ENSMUST00000044524.9
|
Dap
|
death-associated protein |
| chr17_+_3397189 | 0.72 |
ENSMUST00000072156.6
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr17_+_36958623 | 0.71 |
ENSMUST00000173814.1
|
Znrd1as
|
Znrd1 antisense |
| chr5_+_90518932 | 0.71 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr4_-_42661893 | 0.70 |
ENSMUST00000108006.3
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr11_-_49114874 | 0.70 |
ENSMUST00000109201.1
|
Olfr1396
|
olfactory receptor 1396 |
| chrX_+_134187492 | 0.70 |
ENSMUST00000064476.4
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chr1_-_190170671 | 0.69 |
ENSMUST00000175916.1
|
Prox1
|
prospero-related homeobox 1 |
| chr16_-_48771956 | 0.69 |
ENSMUST00000170861.1
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr3_+_86070915 | 0.69 |
ENSMUST00000182666.1
|
Sh3d19
|
SH3 domain protein D19 |
| chr7_+_64501687 | 0.69 |
ENSMUST00000032732.8
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chrX_+_139563316 | 0.69 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr5_+_89028035 | 0.68 |
ENSMUST00000113216.2
ENSMUST00000134303.1 |
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr3_-_92123943 | 0.68 |
ENSMUST00000070284.3
|
Prr9
|
proline rich 9 |
| chr14_-_48665098 | 0.68 |
ENSMUST00000118578.1
|
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chrM_+_7759 | 0.67 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chrM_+_8600 | 0.66 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr5_-_84417359 | 0.66 |
ENSMUST00000113401.1
|
Epha5
|
Eph receptor A5 |
| chr11_-_96075581 | 0.65 |
ENSMUST00000107686.1
ENSMUST00000107684.1 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chr9_+_54980880 | 0.65 |
ENSMUST00000093844.3
|
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr7_-_119895446 | 0.65 |
ENSMUST00000098080.2
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr13_+_93674403 | 0.65 |
ENSMUST00000048001.6
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr6_-_147264124 | 0.64 |
ENSMUST00000052296.6
|
Pthlh
|
parathyroid hormone-like peptide |
| chr11_-_99337930 | 0.63 |
ENSMUST00000100482.2
|
Krt26
|
keratin 26 |
| chr11_-_31671863 | 0.63 |
ENSMUST00000058060.7
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr7_-_90475971 | 0.62 |
ENSMUST00000032843.7
|
Tmem126b
|
transmembrane protein 126B |
| chr7_-_126584220 | 0.62 |
ENSMUST00000128970.1
ENSMUST00000116269.2 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr4_-_83285141 | 0.61 |
ENSMUST00000150522.1
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr1_+_107422681 | 0.60 |
ENSMUST00000112710.1
ENSMUST00000086690.4 |
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7 |
| chr11_+_114851507 | 0.60 |
ENSMUST00000177952.1
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_114851142 | 0.60 |
ENSMUST00000133245.1
ENSMUST00000122967.2 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr12_+_111814170 | 0.60 |
ENSMUST00000021714.7
|
Zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr11_-_100088226 | 0.60 |
ENSMUST00000107419.1
|
Krt32
|
keratin 32 |
| chr2_+_125136692 | 0.58 |
ENSMUST00000099452.2
|
Ctxn2
|
cortexin 2 |
| chr17_-_70853482 | 0.58 |
ENSMUST00000118283.1
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr5_+_89027959 | 0.58 |
ENSMUST00000130041.1
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr11_-_96075655 | 0.57 |
ENSMUST00000090541.5
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chr4_-_14621494 | 0.57 |
ENSMUST00000149633.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_+_63255971 | 0.55 |
ENSMUST00000159561.1
ENSMUST00000095852.3 |
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr5_-_137531204 | 0.55 |
ENSMUST00000150063.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr11_+_23306884 | 0.54 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr7_-_102116705 | 0.54 |
ENSMUST00000084830.1
|
Chrna10
|
cholinergic receptor, nicotinic, alpha polypeptide 10 |
| chr1_-_9298499 | 0.53 |
ENSMUST00000132064.1
|
Sntg1
|
syntrophin, gamma 1 |
| chr3_-_32491969 | 0.53 |
ENSMUST00000164954.1
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr1_+_104768510 | 0.52 |
ENSMUST00000062528.8
|
Cdh20
|
cadherin 20 |
| chr6_-_126645784 | 0.52 |
ENSMUST00000055168.3
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr5_-_137645695 | 0.51 |
ENSMUST00000057314.3
|
Irs3
|
insulin receptor substrate 3 |
| chr8_+_22624019 | 0.50 |
ENSMUST00000033936.6
|
Dkk4
|
dickkopf homolog 4 (Xenopus laevis) |
| chr9_-_106891401 | 0.50 |
ENSMUST00000069036.7
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr16_-_93929512 | 0.50 |
ENSMUST00000177648.1
|
Cldn14
|
claudin 14 |
| chr4_+_43401232 | 0.49 |
ENSMUST00000125399.1
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr4_-_155056784 | 0.49 |
ENSMUST00000131173.2
|
Plch2
|
phospholipase C, eta 2 |
| chr16_+_45094036 | 0.48 |
ENSMUST00000061050.5
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr1_-_190170178 | 0.48 |
ENSMUST00000177288.1
|
Prox1
|
prospero-related homeobox 1 |
| chr8_-_3625274 | 0.48 |
ENSMUST00000004749.6
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr8_-_84662841 | 0.47 |
ENSMUST00000060427.4
|
Ier2
|
immediate early response 2 |
| chr3_-_88461182 | 0.47 |
ENSMUST00000166237.1
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr8_-_67974567 | 0.47 |
ENSMUST00000098696.3
ENSMUST00000038959.9 ENSMUST00000093469.4 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_+_94327984 | 0.47 |
ENSMUST00000107818.2
ENSMUST00000051221.6 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr5_-_66054499 | 0.47 |
ENSMUST00000145625.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr11_-_82890541 | 0.47 |
ENSMUST00000092844.6
ENSMUST00000021033.9 ENSMUST00000018985.8 |
Rad51d
|
RAD51 homolog D |
| chr8_-_61902669 | 0.46 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr11_-_100762928 | 0.46 |
ENSMUST00000107360.2
ENSMUST00000055083.3 |
Hcrt
|
hypocretin |
| chr7_+_64501949 | 0.46 |
ENSMUST00000138829.1
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr9_-_103222063 | 0.46 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chr7_-_73541738 | 0.45 |
ENSMUST00000169922.2
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr18_+_36281069 | 0.45 |
ENSMUST00000051301.3
|
Pura
|
purine rich element binding protein A |
| chr7_+_64502090 | 0.45 |
ENSMUST00000137732.1
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb2_Dlx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.9 | 3.7 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.8 | 2.4 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.6 | 1.8 | GO:1903919 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.6 | 2.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 | 4.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.5 | 2.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.5 | 1.4 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.5 | 1.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.4 | 0.4 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.4 | 1.2 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.4 | 1.2 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.4 | 1.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.4 | 3.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 1.1 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.4 | 1.1 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.3 | 4.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 1.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 2.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 0.8 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.3 | 0.8 | GO:0051795 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.2 | 4.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 0.9 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.2 | 0.6 | GO:0060618 | nipple development(GO:0060618) |
| 0.2 | 0.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 1.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 0.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 0.9 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 0.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 3.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 4.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 1.4 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.2 | 1.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 2.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 0.5 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 1.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.9 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 0.9 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) diencephalon morphogenesis(GO:0048852) |
| 0.2 | 1.8 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 1.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.4 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 0.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.1 | 2.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.7 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 1.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 3.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 1.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.6 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 1.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.8 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.8 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 1.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.1 | 0.3 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.1 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.8 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 4.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.5 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 2.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 2.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.2 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.5 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 1.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.4 | GO:1904936 | forebrain anterior/posterior pattern specification(GO:0021797) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.7 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.1 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.1 | 0.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.6 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.3 | GO:0042364 | cobalamin metabolic process(GO:0009235) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 2.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.7 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 1.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.9 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 1.1 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.9 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 1.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 1.4 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.1 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 1.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:1903671 | cytoplasm organization(GO:0007028) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 | 0.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 2.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 1.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.3 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.2 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.5 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 6.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 2.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 9.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 6.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 2.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 6.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0070695 | HOPS complex(GO:0030897) FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.8 | 2.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.8 | 3.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.7 | 3.7 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.7 | 4.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.5 | 2.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 1.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.4 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 1.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 3.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 1.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 4.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 1.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.7 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.6 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.2 | 5.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 3.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.7 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 1.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.8 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.5 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.3 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 3.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 2.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.8 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 2.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta-activated receptor activity(GO:0005024) transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 3.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 3.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 1.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 3.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |