Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
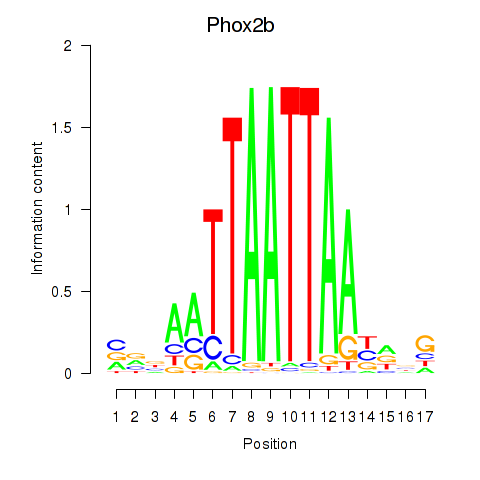
Results for Hoxc4_Arx_Otp_Esx1_Phox2b
Z-value: 0.45
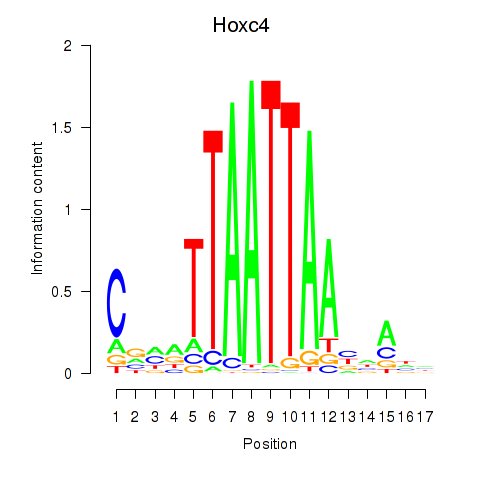
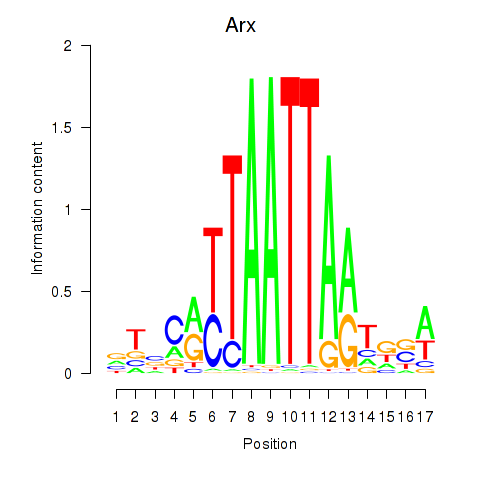
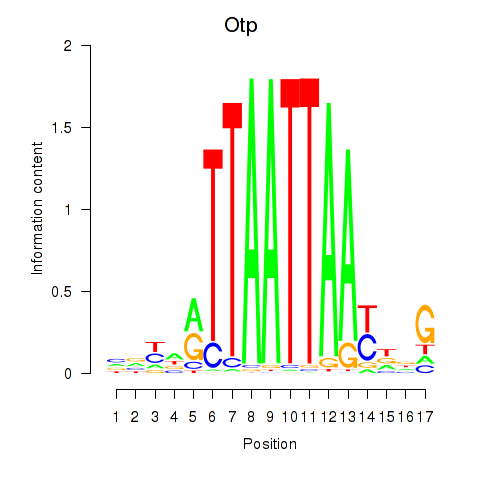
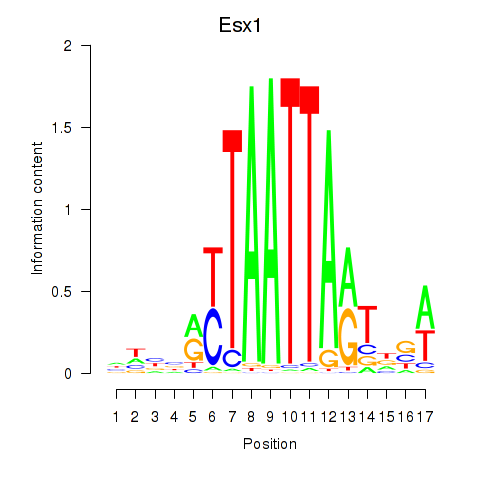
Transcription factors associated with Hoxc4_Arx_Otp_Esx1_Phox2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc4
|
ENSMUSG00000075394.3 | homeobox C4 |
|
Arx
|
ENSMUSG00000035277.9 | aristaless related homeobox |
|
Otp
|
ENSMUSG00000021685.10 | orthopedia homeobox |
|
Esx1
|
ENSMUSG00000023443.7 | extraembryonic, spermatogenesis, homeobox 1 |
|
Phox2b
|
ENSMUSG00000012520.8 | paired-like homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Phox2b | mm10_v2_chr5_-_67099235_67099255 | 0.34 | 4.4e-02 | Click! |
| Esx1 | mm10_v2_chrX_-_137120128_137120171 | 0.23 | 1.8e-01 | Click! |
| Hoxc4 | mm10_v2_chr15_+_103018936_103018973 | 0.13 | 4.6e-01 | Click! |
| Arx | mm10_v2_chrX_+_93286499_93286522 | -0.08 | 6.6e-01 | Click! |
| Otp | mm10_v2_chr13_+_94875600_94875611 | 0.05 | 7.8e-01 | Click! |
Activity profile of Hoxc4_Arx_Otp_Esx1_Phox2b motif
Sorted Z-values of Hoxc4_Arx_Otp_Esx1_Phox2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_160619971 | 2.34 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr4_+_145585166 | 1.65 |
ENSMUST00000105739.1
ENSMUST00000119718.1 |
Gm13212
|
predicted gene 13212 |
| chr4_+_103143052 | 1.51 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr16_+_92478743 | 1.43 |
ENSMUST00000160494.1
|
2410124H12Rik
|
RIKEN cDNA 2410124H12 gene |
| chr10_-_76110956 | 1.12 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr7_+_39588931 | 1.10 |
ENSMUST00000178791.1
ENSMUST00000098511.3 |
Gm2058
|
predicted gene 2058 |
| chr2_-_72813665 | 0.94 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chr7_+_45621805 | 0.89 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chrX_+_9885622 | 0.88 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr13_-_102906046 | 0.84 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_-_102905740 | 0.81 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_55241236 | 0.75 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr17_-_48432723 | 0.75 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr5_-_62765618 | 0.67 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_-_3399545 | 0.64 |
ENSMUST00000120087.3
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr4_+_108719649 | 0.60 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr9_+_119063429 | 0.60 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr10_+_94575257 | 0.58 |
ENSMUST00000121471.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr4_-_147809788 | 0.57 |
ENSMUST00000105734.3
ENSMUST00000176201.1 |
Gm13157
Gm20707
|
predicted gene 13157 predicted gene 20707 |
| chr18_+_4993795 | 0.56 |
ENSMUST00000153016.1
|
Svil
|
supervillin |
| chr8_+_34054622 | 0.55 |
ENSMUST00000149618.1
|
Gm9951
|
predicted gene 9951 |
| chr2_+_174450678 | 0.55 |
ENSMUST00000016399.5
|
Tubb1
|
tubulin, beta 1 class VI |
| chr5_+_13398688 | 0.55 |
ENSMUST00000125629.1
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_-_132075250 | 0.54 |
ENSMUST00000105970.1
ENSMUST00000105975.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr2_-_20943413 | 0.52 |
ENSMUST00000140230.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr5_-_108749448 | 0.52 |
ENSMUST00000068946.7
|
Rnf212
|
ring finger protein 212 |
| chr2_+_119047129 | 0.52 |
ENSMUST00000153300.1
ENSMUST00000028799.5 |
Casc5
|
cancer susceptibility candidate 5 |
| chr2_+_119047116 | 0.50 |
ENSMUST00000152380.1
ENSMUST00000099542.2 |
Casc5
|
cancer susceptibility candidate 5 |
| chr5_-_43981757 | 0.49 |
ENSMUST00000061299.7
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr7_+_18245347 | 0.48 |
ENSMUST00000066780.4
|
Mill1
|
MHC I like leukocyte 1 |
| chr17_+_82539258 | 0.47 |
ENSMUST00000097278.3
|
Gm6594
|
predicted pseudogene 6594 |
| chr4_+_147132038 | 0.47 |
ENSMUST00000084149.3
|
Gm13139
|
predicted gene 13139 |
| chr2_+_3424123 | 0.46 |
ENSMUST00000061852.5
ENSMUST00000100463.3 ENSMUST00000102988.3 ENSMUST00000115066.1 |
Dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr4_+_146097312 | 0.45 |
ENSMUST00000105730.1
ENSMUST00000091878.5 |
Gm13051
|
predicted gene 13051 |
| chr8_-_122915987 | 0.45 |
ENSMUST00000098333.4
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr17_-_78684262 | 0.45 |
ENSMUST00000145480.1
|
Strn
|
striatin, calmodulin binding protein |
| chr4_+_147553277 | 0.40 |
ENSMUST00000139784.1
ENSMUST00000143885.1 ENSMUST00000081742.6 |
Gm13154
|
predicted gene 13154 |
| chr19_+_23723279 | 0.39 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr5_+_90490714 | 0.36 |
ENSMUST00000042755.3
|
Afp
|
alpha fetoprotein |
| chr13_+_120308146 | 0.36 |
ENSMUST00000081558.7
|
BC147527
|
cDNA sequence BC147527 |
| chr14_+_27000362 | 0.35 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr4_+_146971976 | 0.34 |
ENSMUST00000146688.2
|
Gm13150
|
predicted gene 13150 |
| chr9_-_123678873 | 0.34 |
ENSMUST00000040960.6
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr3_+_66219909 | 0.34 |
ENSMUST00000029421.5
|
Ptx3
|
pentraxin related gene |
| chr15_+_98571004 | 0.34 |
ENSMUST00000023728.6
|
4930415O20Rik
|
RIKEN cDNA 4930415O20 gene |
| chr5_-_138170992 | 0.33 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr9_-_20959785 | 0.32 |
ENSMUST00000177754.1
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr2_-_168767136 | 0.31 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr4_+_105789869 | 0.31 |
ENSMUST00000184254.1
|
Gm12728
|
predicted gene 12728 |
| chr10_+_115817247 | 0.31 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr1_+_40439767 | 0.29 |
ENSMUST00000173514.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr7_-_101581161 | 0.28 |
ENSMUST00000063920.2
|
Art2b
|
ADP-ribosyltransferase 2b |
| chr14_-_100149764 | 0.28 |
ENSMUST00000097079.4
|
Klf12
|
Kruppel-like factor 12 |
| chr5_+_37185897 | 0.28 |
ENSMUST00000094840.3
|
Gm1043
|
predicted gene 1043 |
| chrX_-_160906998 | 0.28 |
ENSMUST00000069417.5
|
Gja6
|
gap junction protein, alpha 6 |
| chr4_+_146502027 | 0.27 |
ENSMUST00000105735.2
|
Gm13247
|
predicted gene 13247 |
| chr11_-_49113757 | 0.27 |
ENSMUST00000060398.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr4_+_136172367 | 0.27 |
ENSMUST00000061721.5
|
E2f2
|
E2F transcription factor 2 |
| chr16_+_45224315 | 0.26 |
ENSMUST00000102802.3
ENSMUST00000063654.4 |
Btla
|
B and T lymphocyte associated |
| chr1_+_40439627 | 0.25 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chrX_-_160138375 | 0.24 |
ENSMUST00000033662.8
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr2_-_168767029 | 0.24 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr6_+_37870786 | 0.23 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr5_+_107497718 | 0.23 |
ENSMUST00000112671.2
|
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr6_-_129100903 | 0.23 |
ENSMUST00000032258.6
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr5_-_94306796 | 0.22 |
ENSMUST00000097479.1
|
Gm16522
|
predicted gene, 16522 |
| chr4_-_43558386 | 0.22 |
ENSMUST00000130353.1
|
Tln1
|
talin 1 |
| chr12_-_12940600 | 0.22 |
ENSMUST00000130990.1
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr14_+_58893465 | 0.21 |
ENSMUST00000079960.1
|
Rpl13-ps3
|
ribosomal protein L13, pseudogene 3 |
| chr14_-_118735143 | 0.21 |
ENSMUST00000184172.1
|
RP24-241I19.1
|
RP24-241I19.1 |
| chr9_+_72806874 | 0.21 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr6_-_66559708 | 0.20 |
ENSMUST00000079584.1
|
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr2_-_121235689 | 0.20 |
ENSMUST00000142400.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr17_-_32822200 | 0.20 |
ENSMUST00000179695.1
|
Zfp799
|
zinc finger protein 799 |
| chr13_-_100786402 | 0.20 |
ENSMUST00000174038.1
ENSMUST00000091295.7 ENSMUST00000072119.8 |
Ccnb1
|
cyclin B1 |
| chr18_+_53551594 | 0.20 |
ENSMUST00000115398.1
|
Prdm6
|
PR domain containing 6 |
| chr10_+_37139558 | 0.20 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr3_+_40800778 | 0.19 |
ENSMUST00000169566.1
|
Plk4
|
polo-like kinase 4 |
| chr6_+_123262107 | 0.19 |
ENSMUST00000032240.2
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr11_+_60537978 | 0.18 |
ENSMUST00000044250.3
|
Alkbh5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chrX_+_101449078 | 0.18 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr2_+_150323702 | 0.18 |
ENSMUST00000133235.2
|
Gm10130
|
predicted gene 10130 |
| chr2_+_20737306 | 0.18 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr11_-_107337556 | 0.18 |
ENSMUST00000040380.6
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr10_+_115384951 | 0.18 |
ENSMUST00000036044.8
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr7_+_103937382 | 0.18 |
ENSMUST00000098189.1
|
Olfr632
|
olfactory receptor 632 |
| chr6_-_115037824 | 0.18 |
ENSMUST00000174848.1
ENSMUST00000032461.5 |
Tamm41
|
TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) |
| chr9_-_123678782 | 0.17 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr2_+_69897255 | 0.17 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr18_+_4920509 | 0.17 |
ENSMUST00000126977.1
|
Svil
|
supervillin |
| chr10_-_62379852 | 0.17 |
ENSMUST00000143236.1
ENSMUST00000133429.1 ENSMUST00000132926.1 ENSMUST00000116238.2 |
Hk1
|
hexokinase 1 |
| chr3_-_96263311 | 0.17 |
ENSMUST00000171473.1
|
Hist2h4
|
histone cluster 2, H4 |
| chr8_-_22060019 | 0.16 |
ENSMUST00000110738.2
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr16_-_42340595 | 0.16 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr3_-_17230976 | 0.16 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr16_-_19286837 | 0.16 |
ENSMUST00000056727.4
|
Olfr164
|
olfactory receptor 164 |
| chr5_+_107497762 | 0.16 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr4_+_101986626 | 0.16 |
ENSMUST00000106914.1
|
Gm12789
|
predicted gene 12789 |
| chr12_+_116275386 | 0.16 |
ENSMUST00000090195.4
|
Gm11027
|
predicted gene 11027 |
| chr4_+_102589687 | 0.15 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chrX_-_139871637 | 0.15 |
ENSMUST00000033811.7
ENSMUST00000087401.5 |
Morc4
|
microrchidia 4 |
| chr19_-_32196393 | 0.15 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr1_-_138175126 | 0.15 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_185329331 | 0.15 |
ENSMUST00000027921.4
ENSMUST00000110975.1 ENSMUST00000110974.3 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr4_+_147492417 | 0.15 |
ENSMUST00000105721.2
|
Gm13152
|
predicted gene 13152 |
| chr11_-_87359011 | 0.15 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr4_+_145510759 | 0.15 |
ENSMUST00000105742.1
ENSMUST00000136309.1 |
Gm13225
|
predicted gene 13225 |
| chr13_+_23533869 | 0.15 |
ENSMUST00000073261.2
|
Hist1h2af
|
histone cluster 1, H2af |
| chr3_-_72967854 | 0.15 |
ENSMUST00000167334.1
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr2_-_104493690 | 0.15 |
ENSMUST00000111124.1
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr6_+_92816460 | 0.14 |
ENSMUST00000057977.3
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr4_+_74242468 | 0.14 |
ENSMUST00000077851.3
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr8_-_4779513 | 0.14 |
ENSMUST00000022945.7
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr2_+_130195194 | 0.14 |
ENSMUST00000077988.1
|
Tmc2
|
transmembrane channel-like gene family 2 |
| chr15_-_64922290 | 0.13 |
ENSMUST00000023007.5
|
Adcy8
|
adenylate cyclase 8 |
| chr3_-_113291449 | 0.13 |
ENSMUST00000179568.1
|
Amy2a4
|
amylase 2a4 |
| chr9_-_14381242 | 0.13 |
ENSMUST00000167549.1
|
Endod1
|
endonuclease domain containing 1 |
| chr5_+_3543812 | 0.13 |
ENSMUST00000115527.3
|
Fam133b
|
family with sequence similarity 133, member B |
| chr9_+_32224457 | 0.13 |
ENSMUST00000183121.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr16_+_11406618 | 0.13 |
ENSMUST00000122168.1
|
Snx29
|
sorting nexin 29 |
| chr6_+_34029421 | 0.13 |
ENSMUST00000070189.3
ENSMUST00000101564.2 |
Lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr12_-_118198917 | 0.13 |
ENSMUST00000084806.6
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr15_-_36879816 | 0.13 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr2_-_164585447 | 0.13 |
ENSMUST00000109342.1
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr4_+_146610961 | 0.13 |
ENSMUST00000130825.1
|
Gm13248
|
predicted gene 13248 |
| chr6_-_122340499 | 0.12 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chrX_+_157699113 | 0.12 |
ENSMUST00000112521.1
|
Smpx
|
small muscle protein, X-linked |
| chr10_-_86022325 | 0.12 |
ENSMUST00000181665.1
|
A230060F14Rik
|
RIKEN cDNA A230060F14 gene |
| chr2_-_125782834 | 0.12 |
ENSMUST00000053699.6
|
Secisbp2l
|
SECIS binding protein 2-like |
| chr6_-_118479237 | 0.12 |
ENSMUST00000161170.1
|
Zfp9
|
zinc finger protein 9 |
| chr4_-_136602641 | 0.12 |
ENSMUST00000105847.1
ENSMUST00000116273.2 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr1_-_89933290 | 0.12 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr10_+_116964707 | 0.12 |
ENSMUST00000176050.1
ENSMUST00000176455.1 |
D630029K05Rik
|
RIKEN cDNA D630029K05 gene |
| chr14_-_104522615 | 0.12 |
ENSMUST00000022716.2
|
Rnf219
|
ring finger protein 219 |
| chr10_-_79097560 | 0.12 |
ENSMUST00000039271.6
|
2610008E11Rik
|
RIKEN cDNA 2610008E11 gene |
| chr1_-_4880669 | 0.12 |
ENSMUST00000078030.3
|
Gm6104
|
predicted gene 6104 |
| chr16_-_45724600 | 0.12 |
ENSMUST00000096057.4
|
Tagln3
|
transgelin 3 |
| chr14_+_4514758 | 0.11 |
ENSMUST00000112776.2
|
Gm3173
|
predicted gene 3173 |
| chr17_+_36898110 | 0.11 |
ENSMUST00000078438.4
|
Trim31
|
tripartite motif-containing 31 |
| chr9_+_76014855 | 0.11 |
ENSMUST00000008052.6
ENSMUST00000183425.1 ENSMUST00000183979.1 ENSMUST00000117981.2 |
Hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase-like 1 |
| chr17_-_43543639 | 0.11 |
ENSMUST00000178772.1
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr19_-_47692042 | 0.11 |
ENSMUST00000026045.7
ENSMUST00000086923.5 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr8_-_61902669 | 0.11 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr17_-_33033367 | 0.11 |
ENSMUST00000087654.4
|
Zfp763
|
zinc finger protein 763 |
| chr9_+_38752796 | 0.11 |
ENSMUST00000074740.2
|
Olfr920
|
olfactory receptor 920 |
| chr2_-_131160006 | 0.10 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chrX_+_160768013 | 0.10 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr7_+_29170345 | 0.10 |
ENSMUST00000033886.7
|
Ggn
|
gametogenetin |
| chr10_+_75566257 | 0.10 |
ENSMUST00000129232.1
ENSMUST00000143792.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_+_40354303 | 0.10 |
ENSMUST00000136835.1
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr9_-_39603635 | 0.10 |
ENSMUST00000119722.1
|
AW551984
|
expressed sequence AW551984 |
| chr5_+_91074611 | 0.09 |
ENSMUST00000031324.4
|
Ereg
|
epiregulin |
| chr10_-_117148474 | 0.09 |
ENSMUST00000020381.3
|
Frs2
|
fibroblast growth factor receptor substrate 2 |
| chr19_-_38043559 | 0.09 |
ENSMUST00000041475.8
ENSMUST00000172095.2 |
Myof
|
myoferlin |
| chr2_+_69670100 | 0.09 |
ENSMUST00000100050.3
|
Klhl41
|
kelch-like 41 |
| chr4_-_146909590 | 0.09 |
ENSMUST00000049821.3
|
Gm21411
|
predicted gene, 21411 |
| chrX_+_134686519 | 0.09 |
ENSMUST00000124226.2
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr6_-_128275577 | 0.09 |
ENSMUST00000130454.1
|
Tead4
|
TEA domain family member 4 |
| chr10_-_33951190 | 0.09 |
ENSMUST00000048222.4
|
Zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chr13_-_55100248 | 0.09 |
ENSMUST00000026997.5
ENSMUST00000127195.1 ENSMUST00000099496.3 |
Uimc1
|
ubiquitin interaction motif containing 1 |
| chr19_-_34166037 | 0.09 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr10_+_116143881 | 0.08 |
ENSMUST00000105271.2
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr3_+_28263205 | 0.08 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr8_-_3624989 | 0.08 |
ENSMUST00000142431.1
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr10_+_79996479 | 0.08 |
ENSMUST00000132517.1
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr4_+_146161909 | 0.08 |
ENSMUST00000131932.1
|
Zfp600
|
zinc finger protein 600 |
| chr7_-_25005895 | 0.08 |
ENSMUST00000102858.3
ENSMUST00000080882.6 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr15_+_81744848 | 0.08 |
ENSMUST00000109554.1
|
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr11_-_99244058 | 0.08 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr11_+_51261719 | 0.08 |
ENSMUST00000130641.1
|
Clk4
|
CDC like kinase 4 |
| chr7_+_29170204 | 0.08 |
ENSMUST00000098609.2
|
Ggn
|
gametogenetin |
| chr11_+_121237216 | 0.08 |
ENSMUST00000103015.3
|
Narf
|
nuclear prelamin A recognition factor |
| chr9_-_85749308 | 0.08 |
ENSMUST00000039213.8
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr10_-_88605017 | 0.08 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr11_+_35121126 | 0.07 |
ENSMUST00000069837.3
|
Slit3
|
slit homolog 3 (Drosophila) |
| chr4_+_32623985 | 0.07 |
ENSMUST00000108178.1
|
Casp8ap2
|
caspase 8 associated protein 2 |
| chr1_-_24612700 | 0.07 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr9_+_113812547 | 0.07 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr7_+_128744870 | 0.07 |
ENSMUST00000042942.8
|
Sec23ip
|
Sec23 interacting protein |
| chr5_-_87490869 | 0.07 |
ENSMUST00000147854.1
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr17_-_26099257 | 0.07 |
ENSMUST00000053575.3
|
Gm8186
|
predicted gene 8186 |
| chr2_+_121956651 | 0.07 |
ENSMUST00000110574.1
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chrX_-_23266751 | 0.07 |
ENSMUST00000115316.2
|
Klhl13
|
kelch-like 13 |
| chrX_+_56346390 | 0.07 |
ENSMUST00000101560.3
|
Zfp449
|
zinc finger protein 449 |
| chr15_-_9140374 | 0.07 |
ENSMUST00000096482.3
ENSMUST00000110585.2 |
Skp2
|
S-phase kinase-associated protein 2 (p45) |
| chr9_+_35559460 | 0.07 |
ENSMUST00000034615.3
ENSMUST00000121246.1 |
Pus3
|
pseudouridine synthase 3 |
| chr14_+_26259109 | 0.07 |
ENSMUST00000174494.1
|
Duxbl3
|
double homeobox B-like 3 |
| chr18_-_43477764 | 0.07 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr12_-_103694713 | 0.07 |
ENSMUST00000118101.1
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr1_+_161070767 | 0.07 |
ENSMUST00000111618.1
ENSMUST00000111620.3 ENSMUST00000028035.7 |
Cenpl
|
centromere protein L |
| chr16_+_58408443 | 0.07 |
ENSMUST00000046663.7
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chrX_-_106221145 | 0.07 |
ENSMUST00000113495.2
|
Taf9b
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr18_-_56975333 | 0.06 |
ENSMUST00000139243.2
ENSMUST00000025488.8 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr1_+_63176818 | 0.06 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr14_+_4430992 | 0.06 |
ENSMUST00000164603.1
ENSMUST00000166848.1 |
Gm3173
|
predicted gene 3173 |
| chrX_+_150547375 | 0.06 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr4_-_147702553 | 0.06 |
ENSMUST00000117638.1
|
Zfp534
|
zinc finger protein 534 |
| chr12_-_113361232 | 0.06 |
ENSMUST00000103423.1
|
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr5_+_26817357 | 0.06 |
ENSMUST00000071500.6
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr7_-_68353182 | 0.06 |
ENSMUST00000123509.1
ENSMUST00000129965.1 |
Gm16158
|
predicted gene 16158 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc4_Arx_Otp_Esx1_Phox2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.5 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.3 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.6 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.5 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 1.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.2 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.6 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:2000473 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.0 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:1900113 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) chemorepellent activity(GO:0045499) |
| 0.0 | 1.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |