Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
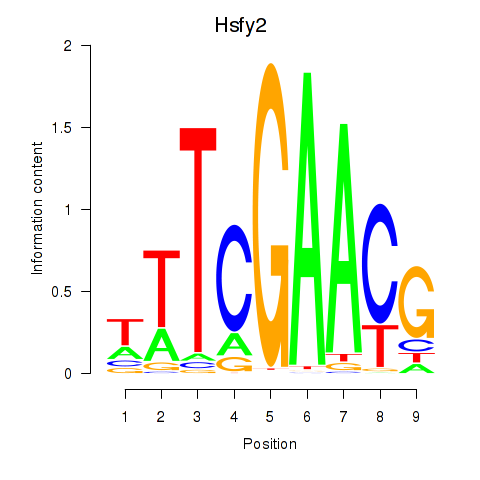
Results for Hsfy2
Z-value: 0.96
Transcription factors associated with Hsfy2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsfy2
|
ENSMUSG00000045336.4 | heat shock transcription factor, Y-linked 2 |
Activity profile of Hsfy2 motif
Sorted Z-values of Hsfy2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_152847993 | 7.00 |
ENSMUST00000028969.8
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr2_+_152847961 | 5.75 |
ENSMUST00000164120.1
ENSMUST00000178997.1 ENSMUST00000109816.1 |
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr11_-_102925086 | 5.13 |
ENSMUST00000021311.9
|
Kif18b
|
kinesin family member 18B |
| chr14_-_67715585 | 5.11 |
ENSMUST00000163100.1
ENSMUST00000132705.1 ENSMUST00000124045.1 |
Cdca2
|
cell division cycle associated 2 |
| chr10_+_20347788 | 4.92 |
ENSMUST00000169712.1
|
Mtfr2
|
mitochondrial fission regulator 2 |
| chr6_+_86628174 | 4.79 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr8_-_53638945 | 4.20 |
ENSMUST00000047768.4
|
Neil3
|
nei like 3 (E. coli) |
| chr11_+_58640394 | 4.06 |
ENSMUST00000075084.4
|
Trim58
|
tripartite motif-containing 58 |
| chr6_+_134929118 | 3.66 |
ENSMUST00000185152.1
ENSMUST00000184504.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr3_-_54915867 | 3.59 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr3_-_84479418 | 3.54 |
ENSMUST00000091002.1
|
Fhdc1
|
FH2 domain containing 1 |
| chr1_+_139454747 | 3.51 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr10_-_79874211 | 3.50 |
ENSMUST00000167897.1
|
BC005764
|
cDNA sequence BC005764 |
| chr11_+_11686213 | 3.33 |
ENSMUST00000076700.4
ENSMUST00000048122.6 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr11_+_32205411 | 3.27 |
ENSMUST00000039601.3
ENSMUST00000149043.1 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr4_+_121039385 | 3.24 |
ENSMUST00000030372.5
|
Col9a2
|
collagen, type IX, alpha 2 |
| chr3_-_20242173 | 3.15 |
ENSMUST00000001921.1
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr7_+_13733502 | 2.91 |
ENSMUST00000086148.6
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr9_-_64172879 | 2.85 |
ENSMUST00000176299.1
ENSMUST00000130127.1 ENSMUST00000176794.1 ENSMUST00000177045.1 |
Zwilch
|
zwilch kinetochore protein |
| chr13_-_37050237 | 2.82 |
ENSMUST00000164727.1
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr10_-_79874233 | 2.69 |
ENSMUST00000166023.1
ENSMUST00000167707.1 ENSMUST00000165601.1 |
BC005764
|
cDNA sequence BC005764 |
| chr4_+_126556935 | 2.58 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr3_+_107896247 | 2.55 |
ENSMUST00000169365.1
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr8_-_22185758 | 2.52 |
ENSMUST00000046916.7
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr7_-_126704522 | 2.43 |
ENSMUST00000135087.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr3_+_108383829 | 2.40 |
ENSMUST00000090561.3
ENSMUST00000102629.1 ENSMUST00000128089.1 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr9_-_39603635 | 2.29 |
ENSMUST00000119722.1
|
AW551984
|
expressed sequence AW551984 |
| chr7_-_127993831 | 2.25 |
ENSMUST00000033056.3
|
Pycard
|
PYD and CARD domain containing |
| chr1_-_93342734 | 2.20 |
ENSMUST00000027493.3
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr4_+_126556994 | 2.06 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr17_+_47593516 | 2.05 |
ENSMUST00000182874.1
|
Ccnd3
|
cyclin D3 |
| chr14_-_31577318 | 2.01 |
ENSMUST00000112027.2
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr9_-_72491939 | 2.00 |
ENSMUST00000185151.1
ENSMUST00000085358.5 ENSMUST00000184125.1 ENSMUST00000183574.1 ENSMUST00000184831.1 |
Tex9
|
testis expressed gene 9 |
| chr15_+_44457522 | 1.97 |
ENSMUST00000166957.1
ENSMUST00000038336.5 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr11_+_5569679 | 1.95 |
ENSMUST00000109856.1
ENSMUST00000109855.1 ENSMUST00000118112.2 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr1_+_191063001 | 1.94 |
ENSMUST00000076952.5
ENSMUST00000139340.1 ENSMUST00000078259.6 |
Nsl1
|
NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) |
| chr6_+_123229843 | 1.94 |
ENSMUST00000112554.2
ENSMUST00000024118.4 ENSMUST00000117130.1 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr3_-_86920830 | 1.94 |
ENSMUST00000029719.8
|
Dclk2
|
doublecortin-like kinase 2 |
| chr17_+_47593444 | 1.90 |
ENSMUST00000182209.1
|
Ccnd3
|
cyclin D3 |
| chr9_+_106281061 | 1.84 |
ENSMUST00000072206.6
|
Poc1a
|
POC1 centriolar protein homolog A (Chlamydomonas) |
| chr10_+_45577811 | 1.81 |
ENSMUST00000037044.6
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr15_-_80083374 | 1.78 |
ENSMUST00000081650.7
|
Rpl3
|
ribosomal protein L3 |
| chr9_-_36726374 | 1.75 |
ENSMUST00000172702.2
ENSMUST00000172742.1 ENSMUST00000034625.5 |
Chek1
|
checkpoint kinase 1 |
| chr17_+_26917091 | 1.74 |
ENSMUST00000078961.4
|
Kifc5b
|
kinesin family member C5B |
| chr7_+_43351378 | 1.73 |
ENSMUST00000012798.7
ENSMUST00000122423.1 ENSMUST00000121494.1 |
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr6_+_48589445 | 1.65 |
ENSMUST00000064744.3
|
Gm5111
|
predicted gene 5111 |
| chr9_-_32344237 | 1.65 |
ENSMUST00000034533.5
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr14_+_79451791 | 1.63 |
ENSMUST00000100359.1
|
Zbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr7_-_14123042 | 1.59 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr12_-_55014329 | 1.58 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr10_+_79664553 | 1.57 |
ENSMUST00000020554.6
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr13_-_95525239 | 1.53 |
ENSMUST00000022185.8
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr1_-_190978954 | 1.48 |
ENSMUST00000047409.6
|
Vash2
|
vasohibin 2 |
| chr15_+_103240405 | 1.47 |
ENSMUST00000036004.9
ENSMUST00000087351.7 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr3_+_129878571 | 1.47 |
ENSMUST00000029629.8
|
Pla2g12a
|
phospholipase A2, group XIIA |
| chr15_-_66801577 | 1.43 |
ENSMUST00000168589.1
|
Sla
|
src-like adaptor |
| chr13_+_5861489 | 1.43 |
ENSMUST00000000080.6
|
Klf6
|
Kruppel-like factor 6 |
| chr6_+_86404257 | 1.40 |
ENSMUST00000095752.2
ENSMUST00000130967.1 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr14_+_60378242 | 1.38 |
ENSMUST00000022561.6
|
Amer2
|
APC membrane recruitment 2 |
| chrX_+_94636066 | 1.37 |
ENSMUST00000096368.3
|
Gspt2
|
G1 to S phase transition 2 |
| chr15_+_38933142 | 1.37 |
ENSMUST00000163313.1
|
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr5_+_108132885 | 1.36 |
ENSMUST00000047677.7
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr6_+_86404219 | 1.36 |
ENSMUST00000095754.3
ENSMUST00000095753.2 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chr1_-_190979280 | 1.35 |
ENSMUST00000166139.1
|
Vash2
|
vasohibin 2 |
| chr13_-_120027011 | 1.35 |
ENSMUST00000177659.1
|
Gm21370
|
predicted gene, 21370 |
| chr2_+_172393900 | 1.33 |
ENSMUST00000109136.2
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr1_-_33669745 | 1.33 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr4_+_108879130 | 1.33 |
ENSMUST00000106651.2
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr2_-_181693810 | 1.32 |
ENSMUST00000108776.1
ENSMUST00000108771.1 ENSMUST00000108779.1 ENSMUST00000108769.1 ENSMUST00000108772.1 ENSMUST00000002532.2 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr11_+_102881204 | 1.31 |
ENSMUST00000021307.3
ENSMUST00000159834.1 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr4_+_108879063 | 1.30 |
ENSMUST00000106650.2
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr11_-_11462408 | 1.29 |
ENSMUST00000020413.3
|
Zpbp
|
zona pellucida binding protein |
| chr7_-_112159034 | 1.28 |
ENSMUST00000033036.5
|
Dkk3
|
dickkopf homolog 3 (Xenopus laevis) |
| chr2_+_119799514 | 1.26 |
ENSMUST00000028763.9
|
Tyro3
|
TYRO3 protein tyrosine kinase 3 |
| chr8_+_60655540 | 1.25 |
ENSMUST00000034066.3
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr6_+_49367739 | 1.24 |
ENSMUST00000060561.8
ENSMUST00000121903.1 ENSMUST00000134786.1 |
Fam221a
|
family with sequence similarity 221, member A |
| chrX_+_162901567 | 1.21 |
ENSMUST00000112303.1
ENSMUST00000033727.7 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr9_+_64173364 | 1.21 |
ENSMUST00000034966.7
|
Rpl4
|
ribosomal protein L4 |
| chr11_+_62248977 | 1.19 |
ENSMUST00000018644.2
|
Adora2b
|
adenosine A2b receptor |
| chr3_-_152340350 | 1.19 |
ENSMUST00000073089.6
ENSMUST00000068243.6 |
Fam73a
|
family with sequence similarity 73, member A |
| chr19_-_11660516 | 1.18 |
ENSMUST00000135994.1
ENSMUST00000121793.1 ENSMUST00000069681.3 |
Plac1l
|
placenta-specific 1-like |
| chrX_+_159627265 | 1.17 |
ENSMUST00000112456.2
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr6_-_40951826 | 1.17 |
ENSMUST00000073642.5
|
Gm4744
|
predicted gene 4744 |
| chrX_-_133688978 | 1.17 |
ENSMUST00000149154.1
ENSMUST00000167944.1 |
Pcdh19
|
protocadherin 19 |
| chr5_-_149051604 | 1.16 |
ENSMUST00000093196.4
|
Hmgb1
|
high mobility group box 1 |
| chr17_-_14694223 | 1.15 |
ENSMUST00000170872.1
|
Thbs2
|
thrombospondin 2 |
| chr11_-_109828021 | 1.15 |
ENSMUST00000020941.4
|
1700012B07Rik
|
RIKEN cDNA 1700012B07 gene |
| chr5_+_31697960 | 1.15 |
ENSMUST00000114515.2
|
Bre
|
brain and reproductive organ-expressed protein |
| chr7_+_28692849 | 1.15 |
ENSMUST00000039998.4
|
Fbxo27
|
F-box protein 27 |
| chr19_+_18713192 | 1.14 |
ENSMUST00000062753.2
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene |
| chr13_+_55445301 | 1.14 |
ENSMUST00000001115.8
ENSMUST00000099482.3 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr13_+_44729535 | 1.14 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_+_28693032 | 1.13 |
ENSMUST00000151227.1
ENSMUST00000108281.1 |
Fbxo27
|
F-box protein 27 |
| chr11_+_32205483 | 1.10 |
ENSMUST00000121182.1
|
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr4_-_149126688 | 1.08 |
ENSMUST00000030815.2
|
Cort
|
cortistatin |
| chr5_-_113015473 | 1.07 |
ENSMUST00000065167.4
|
Adrbk2
|
adrenergic receptor kinase, beta 2 |
| chrX_+_159840463 | 1.06 |
ENSMUST00000112451.1
ENSMUST00000112453.2 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_-_40733373 | 1.06 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr14_-_99099701 | 1.05 |
ENSMUST00000042471.9
|
Dis3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr7_+_28440927 | 1.04 |
ENSMUST00000078845.6
|
Gmfg
|
glia maturation factor, gamma |
| chr17_+_50698525 | 1.03 |
ENSMUST00000061681.7
|
Gm7334
|
predicted gene 7334 |
| chr5_-_31241215 | 1.02 |
ENSMUST00000068997.3
|
Gm9970
|
predicted gene 9970 |
| chr5_+_31698050 | 1.01 |
ENSMUST00000114507.3
ENSMUST00000063813.4 ENSMUST00000071531.5 ENSMUST00000131995.1 |
Bre
|
brain and reproductive organ-expressed protein |
| chr1_-_149961230 | 1.00 |
ENSMUST00000070200.8
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr7_+_28441026 | 1.00 |
ENSMUST00000135686.1
|
Gmfg
|
glia maturation factor, gamma |
| chr9_-_44344159 | 1.00 |
ENSMUST00000077353.7
|
Hmbs
|
hydroxymethylbilane synthase |
| chr9_-_41004599 | 0.99 |
ENSMUST00000180384.1
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr11_+_3330401 | 0.98 |
ENSMUST00000045153.4
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr9_-_41004562 | 0.96 |
ENSMUST00000034519.6
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr19_+_18713225 | 0.94 |
ENSMUST00000055792.7
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene |
| chr18_-_37644185 | 0.93 |
ENSMUST00000066272.4
|
Taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr7_+_29170345 | 0.92 |
ENSMUST00000033886.7
|
Ggn
|
gametogenetin |
| chr10_-_80918212 | 0.92 |
ENSMUST00000057623.7
ENSMUST00000179022.1 |
Lmnb2
|
lamin B2 |
| chr13_-_105271039 | 0.91 |
ENSMUST00000069686.6
|
Rnf180
|
ring finger protein 180 |
| chr4_-_140845770 | 0.89 |
ENSMUST00000026378.3
|
Padi1
|
peptidyl arginine deiminase, type I |
| chr11_+_3330781 | 0.89 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_-_51396716 | 0.89 |
ENSMUST00000141156.1
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr6_-_56362356 | 0.89 |
ENSMUST00000044505.7
ENSMUST00000166102.1 ENSMUST00000164037.1 ENSMUST00000114327.2 |
Pde1c
|
phosphodiesterase 1C |
| chr4_-_41464816 | 0.89 |
ENSMUST00000108055.2
ENSMUST00000154535.1 ENSMUST00000030148.5 |
Kif24
|
kinesin family member 24 |
| chr11_+_44518959 | 0.87 |
ENSMUST00000019333.3
|
Rnf145
|
ring finger protein 145 |
| chr15_+_92344359 | 0.87 |
ENSMUST00000181901.1
|
Gm26760
|
predicted gene, 26760 |
| chr9_-_106096776 | 0.87 |
ENSMUST00000121963.1
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr4_+_119195353 | 0.86 |
ENSMUST00000106345.2
|
Ccdc23
|
coiled-coil domain containing 23 |
| chr17_-_33685386 | 0.85 |
ENSMUST00000139302.1
ENSMUST00000087582.5 ENSMUST00000114385.2 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr9_-_49798729 | 0.85 |
ENSMUST00000166811.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr8_+_94214567 | 0.85 |
ENSMUST00000079961.6
|
Nup93
|
nucleoporin 93 |
| chr8_-_4325096 | 0.83 |
ENSMUST00000098950.4
|
Elavl1
|
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| chr9_-_37613715 | 0.81 |
ENSMUST00000002013.9
|
Spa17
|
sperm autoantigenic protein 17 |
| chr3_-_15838643 | 0.81 |
ENSMUST00000148194.1
|
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr7_+_3704025 | 0.81 |
ENSMUST00000108623.1
ENSMUST00000139818.1 ENSMUST00000108625.1 |
Rps9
|
ribosomal protein S9 |
| chr13_+_23752267 | 0.80 |
ENSMUST00000091703.2
|
Hist1h3b
|
histone cluster 1, H3b |
| chr9_-_45984816 | 0.80 |
ENSMUST00000172450.1
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr7_-_14562171 | 0.78 |
ENSMUST00000181796.1
|
Vmn1r90
|
vomeronasal 1 receptor 90 |
| chr8_-_84840627 | 0.78 |
ENSMUST00000003911.6
ENSMUST00000109761.2 ENSMUST00000128035.1 |
Rad23a
|
RAD23a homolog (S. cerevisiae) |
| chr1_-_21961581 | 0.77 |
ENSMUST00000029667.6
ENSMUST00000173058.1 ENSMUST00000173404.1 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr13_+_51645232 | 0.77 |
ENSMUST00000075853.5
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr19_+_46396885 | 0.76 |
ENSMUST00000039922.6
ENSMUST00000111867.2 ENSMUST00000120778.1 |
Sufu
|
suppressor of fused homolog (Drosophila) |
| chr11_-_102880925 | 0.76 |
ENSMUST00000021306.7
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr10_-_13324160 | 0.75 |
ENSMUST00000105545.4
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr12_-_110845184 | 0.75 |
ENSMUST00000180988.1
|
Gm26912
|
predicted gene, 26912 |
| chr17_+_35241838 | 0.75 |
ENSMUST00000173731.1
|
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr3_-_59220150 | 0.75 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr11_-_20332654 | 0.74 |
ENSMUST00000004634.6
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr9_+_95857597 | 0.74 |
ENSMUST00000034980.7
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr14_-_54781886 | 0.73 |
ENSMUST00000022787.6
|
Slc7a8
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 |
| chr7_+_60155538 | 0.73 |
ENSMUST00000057611.4
|
Gm7367
|
predicted pseudogene 7367 |
| chr8_+_60632856 | 0.73 |
ENSMUST00000160719.1
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr1_-_93801840 | 0.73 |
ENSMUST00000112890.2
ENSMUST00000027503.7 |
Dtymk
|
deoxythymidylate kinase |
| chr4_+_3940747 | 0.72 |
ENSMUST00000119403.1
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chrX_+_101274198 | 0.71 |
ENSMUST00000117203.1
ENSMUST00000087948.4 ENSMUST00000087956.5 |
Med12
|
mediator of RNA polymerase II transcription, subunit 12 homolog (yeast) |
| chr9_-_83146601 | 0.71 |
ENSMUST00000162246.2
ENSMUST00000161796.2 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr14_+_56402656 | 0.71 |
ENSMUST00000095793.1
|
Rnf17
|
ring finger protein 17 |
| chr9_-_107289847 | 0.70 |
ENSMUST00000035194.2
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr2_-_151980135 | 0.70 |
ENSMUST00000062047.5
|
Fam110a
|
family with sequence similarity 110, member A |
| chr5_-_123721007 | 0.68 |
ENSMUST00000031376.7
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chrX_+_101274023 | 0.67 |
ENSMUST00000117706.1
|
Med12
|
mediator of RNA polymerase II transcription, subunit 12 homolog (yeast) |
| chr17_-_31129602 | 0.65 |
ENSMUST00000024827.4
|
Tff3
|
trefoil factor 3, intestinal |
| chr1_+_75546258 | 0.65 |
ENSMUST00000124341.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr4_+_5644084 | 0.65 |
ENSMUST00000054857.6
|
Fam110b
|
family with sequence similarity 110, member B |
| chr11_-_102880981 | 0.65 |
ENSMUST00000107060.1
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr7_+_3703979 | 0.64 |
ENSMUST00000006496.8
|
Rps9
|
ribosomal protein S9 |
| chrX_-_73824938 | 0.64 |
ENSMUST00000114438.2
ENSMUST00000002080.5 |
Pdzd4
|
PDZ domain containing 4 |
| chr6_-_83441674 | 0.64 |
ENSMUST00000089622.4
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr5_+_31697665 | 0.64 |
ENSMUST00000080598.7
|
Bre
|
brain and reproductive organ-expressed protein |
| chr17_+_35241746 | 0.63 |
ENSMUST00000068056.5
ENSMUST00000174757.1 |
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr7_+_3704307 | 0.63 |
ENSMUST00000108624.1
ENSMUST00000126562.1 |
Rps9
|
ribosomal protein S9 |
| chr7_+_128246812 | 0.63 |
ENSMUST00000164710.1
ENSMUST00000070656.5 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr8_+_84872105 | 0.63 |
ENSMUST00000136026.1
ENSMUST00000170296.1 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr16_+_31933851 | 0.62 |
ENSMUST00000052174.2
|
Pigz
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr10_-_7663245 | 0.62 |
ENSMUST00000163085.1
ENSMUST00000159917.1 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr9_+_44134562 | 0.61 |
ENSMUST00000034650.8
ENSMUST00000098852.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr3_-_61365951 | 0.61 |
ENSMUST00000066298.2
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr15_-_32244632 | 0.60 |
ENSMUST00000181536.1
|
0610007N19Rik
|
RIKEN cDNA 0610007N19 |
| chr6_+_129397478 | 0.60 |
ENSMUST00000112081.2
ENSMUST00000112079.2 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr1_-_85254548 | 0.60 |
ENSMUST00000161685.1
|
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chrX_-_57393020 | 0.60 |
ENSMUST00000143310.1
ENSMUST00000098470.2 ENSMUST00000114726.1 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr9_+_88581036 | 0.60 |
ENSMUST00000164661.2
|
Trim43a
|
tripartite motif-containing 43A |
| chr11_-_70410010 | 0.59 |
ENSMUST00000019065.3
ENSMUST00000135148.1 |
Pelp1
|
proline, glutamic acid and leucine rich protein 1 |
| chr17_-_24205514 | 0.59 |
ENSMUST00000097376.3
ENSMUST00000168410.2 ENSMUST00000167791.2 ENSMUST00000040474.7 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr5_+_137629169 | 0.58 |
ENSMUST00000176667.1
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr14_+_75016027 | 0.58 |
ENSMUST00000036072.7
|
5031414D18Rik
|
RIKEN cDNA 5031414D18 gene |
| chr6_-_92244645 | 0.57 |
ENSMUST00000006046.4
|
Trh
|
thyrotropin releasing hormone |
| chr8_+_60632818 | 0.57 |
ENSMUST00000161421.1
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr1_+_44551483 | 0.57 |
ENSMUST00000074525.3
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr18_-_61259987 | 0.56 |
ENSMUST00000170335.2
|
Rps2-ps10
|
ribosomal protein S2, pseudogene 10 |
| chr16_-_64479148 | 0.56 |
ENSMUST00000089279.4
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr12_+_99964499 | 0.56 |
ENSMUST00000177549.1
ENSMUST00000160413.1 ENSMUST00000162221.1 ENSMUST00000049788.8 |
Kcnk13
|
potassium channel, subfamily K, member 13 |
| chr12_-_102743625 | 0.56 |
ENSMUST00000173760.2
ENSMUST00000178384.1 |
Moap1
|
modulator of apoptosis 1 |
| chr8_-_35495487 | 0.55 |
ENSMUST00000033927.6
|
Eri1
|
exoribonuclease 1 |
| chr2_+_25180737 | 0.55 |
ENSMUST00000104999.2
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr14_+_70100083 | 0.55 |
ENSMUST00000022680.7
|
Bin3
|
bridging integrator 3 |
| chr16_+_13780699 | 0.54 |
ENSMUST00000023363.6
|
Rrn3
|
RRN3 RNA polymerase I transcription factor homolog (yeast) |
| chr2_-_154892887 | 0.53 |
ENSMUST00000099173.4
|
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
| chr3_+_122924353 | 0.52 |
ENSMUST00000106429.3
|
1810037I17Rik
|
RIKEN cDNA 1810037I17 gene |
| chr12_+_38781093 | 0.51 |
ENSMUST00000161513.1
|
Etv1
|
ets variant gene 1 |
| chr6_-_129717132 | 0.51 |
ENSMUST00000088046.1
|
Klri1
|
killer cell lectin-like receptor family I member 1 |
| chr10_+_7667503 | 0.51 |
ENSMUST00000040135.8
|
Nup43
|
nucleoporin 43 |
| chr3_+_14533817 | 0.50 |
ENSMUST00000169079.1
ENSMUST00000091325.3 |
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr15_+_82016420 | 0.50 |
ENSMUST00000168581.1
ENSMUST00000170630.1 ENSMUST00000164779.1 |
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr2_-_154892782 | 0.49 |
ENSMUST00000166171.1
ENSMUST00000161172.1 |
Eif2s2
|
eukaryotic translation initiation factor 2, subunit 2 (beta) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsfy2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.8 | 2.3 | GO:0002582 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.7 | 2.0 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.7 | 2.6 | GO:0097494 | positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of vesicle size(GO:0097494) |
| 0.6 | 1.9 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.6 | 3.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.5 | 1.5 | GO:1900135 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.5 | 1.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.5 | 1.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 4.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 15.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.4 | 2.4 | GO:0032796 | uropod organization(GO:0032796) |
| 0.4 | 1.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.4 | 2.5 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.3 | 2.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.3 | 1.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.8 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 1.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.3 | 0.9 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.3 | 2.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 1.9 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 1.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.3 | 1.3 | GO:0032345 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.3 | 1.8 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.2 | 0.7 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 | 0.7 | GO:0009197 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.2 | 1.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.2 | 2.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.2 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.2 | 2.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 1.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.7 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.7 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 0.5 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.2 | 4.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 0.9 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 0.5 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 2.3 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 | 0.9 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 1.0 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.5 | GO:2000373 | CRD-mediated mRNA stabilization(GO:0070934) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.2 | 1.3 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.2 | 1.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 1.0 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.2 | 0.8 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.7 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.4 | GO:1990523 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) negative regulation of sensory perception of pain(GO:1904057) bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0043096 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) purine nucleobase salvage(GO:0043096) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 0.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 8.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.4 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.4 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.6 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.9 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.1 | 0.5 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.3 | GO:0060068 | vagina development(GO:0060068) |
| 0.1 | 4.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 1.2 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 1.0 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 1.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 2.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 1.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.5 | GO:0046813 | T cell antigen processing and presentation(GO:0002457) receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.3 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 1.2 | GO:0036093 | germ cell proliferation(GO:0036093) |
| 0.1 | 4.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.6 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.1 | 4.3 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.9 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.5 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.1 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 1.5 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.6 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.6 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.9 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.6 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 2.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.3 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.4 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 1.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.8 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.6 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.5 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 1.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 1.2 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 1.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 3.2 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 1.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.1 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.9 | GO:0005818 | aster(GO:0005818) |
| 1.0 | 2.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.9 | 3.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.8 | 3.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.6 | 2.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.5 | 1.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 1.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 2.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 1.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 1.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 3.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 3.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 0.8 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 4.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.8 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 1.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 5.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 2.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 1.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 2.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.7 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.8 | 4.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.8 | 4.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.6 | 1.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.5 | 1.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 1.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.4 | 1.2 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.4 | 1.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 2.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 1.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 1.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 0.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 2.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.7 | GO:0050145 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.2 | 0.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 3.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.6 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.2 | 1.6 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.2 | 0.6 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.6 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 0.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 1.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 1.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 4.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 1.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 2.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.9 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.7 | GO:0032405 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.1 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 3.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 3.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 3.3 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 2.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 2.5 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 4.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 8.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0080084 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.1 | 0.7 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 2.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:1902282 | ATP-activated inward rectifier potassium channel activity(GO:0015272) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 1.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 1.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 5.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 4.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 4.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 4.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 2.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 3.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 3.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |