Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
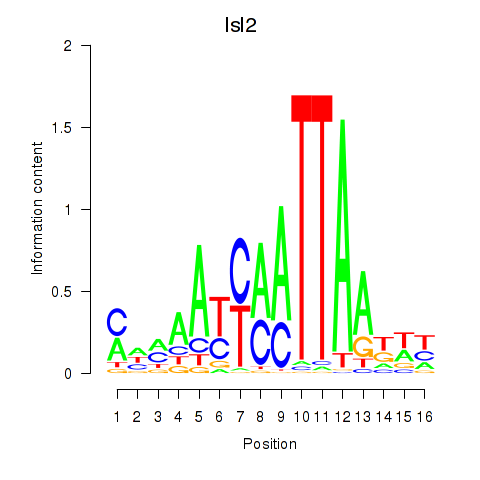
Results for Isl2
Z-value: 1.18
Transcription factors associated with Isl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Isl2
|
ENSMUSG00000032318.6 | insulin related protein 2 (islet 2) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl2 | mm10_v2_chr9_+_55541148_55541209 | -0.69 | 2.6e-06 | Click! |
Activity profile of Isl2 motif
Sorted Z-values of Isl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_8131982 | 15.88 |
ENSMUST00000065651.4
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr9_-_48605147 | 14.37 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr1_-_139858684 | 9.82 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr19_-_7966000 | 9.45 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr8_+_114133601 | 9.27 |
ENSMUST00000109109.1
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr4_+_115299046 | 8.89 |
ENSMUST00000084343.3
|
Cyp4a12a
|
cytochrome P450, family 4, subfamily a, polypeptide 12a |
| chr3_+_138374121 | 8.84 |
ENSMUST00000171054.1
|
Adh6-ps1
|
alcohol dehydrogenase 6 (class V), pseudogene 1 |
| chr8_+_114133635 | 8.31 |
ENSMUST00000147605.1
ENSMUST00000134593.1 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_+_114133557 | 7.51 |
ENSMUST00000073521.5
ENSMUST00000066514.6 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr19_+_39510844 | 6.85 |
ENSMUST00000025968.4
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr3_-_67515487 | 6.17 |
ENSMUST00000178314.1
ENSMUST00000054825.4 |
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr14_-_64455903 | 5.43 |
ENSMUST00000067927.7
|
Msra
|
methionine sulfoxide reductase A |
| chr1_-_121332545 | 5.12 |
ENSMUST00000161068.1
|
Insig2
|
insulin induced gene 2 |
| chr19_+_12695783 | 4.89 |
ENSMUST00000025598.3
ENSMUST00000138545.1 ENSMUST00000154822.1 |
Keg1
|
kidney expressed gene 1 |
| chr19_-_39649046 | 4.87 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr7_-_100656953 | 4.73 |
ENSMUST00000107046.1
ENSMUST00000107045.1 ENSMUST00000139708.1 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr15_-_3303521 | 4.72 |
ENSMUST00000165386.1
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr1_+_88211956 | 4.40 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr1_-_121332571 | 4.23 |
ENSMUST00000071064.6
|
Insig2
|
insulin induced gene 2 |
| chr9_+_119102463 | 4.05 |
ENSMUST00000140326.1
ENSMUST00000165231.1 |
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr10_-_31445921 | 3.91 |
ENSMUST00000000305.5
|
Tpd52l1
|
tumor protein D52-like 1 |
| chr17_-_56005566 | 3.86 |
ENSMUST00000043785.6
|
Stap2
|
signal transducing adaptor family member 2 |
| chr3_-_113574242 | 3.80 |
ENSMUST00000142505.2
|
Amy1
|
amylase 1, salivary |
| chr9_-_15301555 | 3.79 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr17_+_56005672 | 3.49 |
ENSMUST00000133998.1
|
Mpnd
|
MPN domain containing |
| chr5_+_87000838 | 3.39 |
ENSMUST00000031186.7
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr1_+_88055377 | 3.15 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_88070765 | 3.10 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr9_-_51328898 | 3.06 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr1_+_88055467 | 3.05 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr14_-_68533689 | 3.04 |
ENSMUST00000022640.7
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr13_-_12461432 | 2.86 |
ENSMUST00000143693.1
ENSMUST00000144283.1 ENSMUST00000099820.3 ENSMUST00000135166.1 |
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr7_+_119617804 | 2.85 |
ENSMUST00000135683.1
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr5_+_90561102 | 2.78 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr4_+_102589687 | 2.73 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr19_+_45076105 | 2.70 |
ENSMUST00000026234.4
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chrM_+_14138 | 2.61 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr13_+_67833235 | 2.59 |
ENSMUST00000060609.7
|
Gm10037
|
predicted gene 10037 |
| chr2_+_69135799 | 2.48 |
ENSMUST00000041865.7
|
Nostrin
|
nitric oxide synthase trafficker |
| chrX_+_101383726 | 2.47 |
ENSMUST00000119190.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr4_+_43493345 | 2.43 |
ENSMUST00000030181.5
ENSMUST00000107922.2 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr13_+_93674403 | 2.36 |
ENSMUST00000048001.6
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr1_-_187215454 | 2.35 |
ENSMUST00000183819.1
|
Spata17
|
spermatogenesis associated 17 |
| chr2_+_109917639 | 2.29 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr11_+_109543694 | 2.28 |
ENSMUST00000106696.1
|
Arsg
|
arylsulfatase G |
| chr16_+_11406618 | 2.22 |
ENSMUST00000122168.1
|
Snx29
|
sorting nexin 29 |
| chr11_-_49113757 | 2.19 |
ENSMUST00000060398.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr4_-_40722307 | 2.19 |
ENSMUST00000181475.1
|
Gm6297
|
predicted gene 6297 |
| chr10_+_75037291 | 2.18 |
ENSMUST00000139384.1
|
Rab36
|
RAB36, member RAS oncogene family |
| chrM_+_10167 | 2.18 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_-_187215421 | 2.17 |
ENSMUST00000110945.3
ENSMUST00000183931.1 ENSMUST00000027908.6 |
Spata17
|
spermatogenesis associated 17 |
| chrM_-_14060 | 2.17 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr17_+_85028347 | 2.16 |
ENSMUST00000024944.7
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr2_-_110305730 | 2.11 |
ENSMUST00000046233.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chrM_+_9452 | 2.10 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr9_+_72958785 | 2.07 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chrM_+_9870 | 2.04 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_-_79841729 | 2.03 |
ENSMUST00000168038.1
|
Tmem144
|
transmembrane protein 144 |
| chr10_-_77166545 | 2.01 |
ENSMUST00000081654.6
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr6_-_130231638 | 1.98 |
ENSMUST00000088011.4
ENSMUST00000112013.1 ENSMUST00000049304.7 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr2_+_69897220 | 1.90 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr17_-_40319205 | 1.85 |
ENSMUST00000026498.4
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr4_+_22357543 | 1.84 |
ENSMUST00000039234.3
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr11_+_5520652 | 1.83 |
ENSMUST00000063084.9
|
Xbp1
|
X-box binding protein 1 |
| chr19_-_39812744 | 1.80 |
ENSMUST00000162507.1
ENSMUST00000160476.1 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chrM_+_3906 | 1.79 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_101278927 | 1.73 |
ENSMUST00000168089.1
ENSMUST00000017332.3 |
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chr6_-_83121385 | 1.73 |
ENSMUST00000146328.1
ENSMUST00000113936.3 ENSMUST00000032111.4 |
Wbp1
|
WW domain binding protein 1 |
| chr3_+_89459118 | 1.65 |
ENSMUST00000029564.5
|
Pmvk
|
phosphomevalonate kinase |
| chr11_+_103649498 | 1.65 |
ENSMUST00000057870.2
|
Rprml
|
reprimo-like |
| chr9_+_108290433 | 1.62 |
ENSMUST00000035227.6
|
Nicn1
|
nicolin 1 |
| chr5_-_88675190 | 1.61 |
ENSMUST00000133532.1
ENSMUST00000150438.1 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr1_-_24612700 | 1.60 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr6_+_41302265 | 1.57 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr5_-_3647806 | 1.56 |
ENSMUST00000119783.1
ENSMUST00000007559.8 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chrX_-_60893430 | 1.53 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr12_-_84617326 | 1.51 |
ENSMUST00000021666.4
|
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr5_-_87490869 | 1.50 |
ENSMUST00000147854.1
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr6_-_115592571 | 1.48 |
ENSMUST00000112957.1
|
2510049J12Rik
|
RIKEN cDNA 2510049J12 gene |
| chrX_-_103981242 | 1.47 |
ENSMUST00000121153.1
ENSMUST00000070705.4 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr3_+_87435820 | 1.47 |
ENSMUST00000178261.1
ENSMUST00000049926.8 ENSMUST00000166297.1 |
Fcrl5
|
Fc receptor-like 5 |
| chr19_-_39886730 | 1.45 |
ENSMUST00000168838.1
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr11_+_94327984 | 1.43 |
ENSMUST00000107818.2
ENSMUST00000051221.6 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr9_+_119341487 | 1.43 |
ENSMUST00000175743.1
ENSMUST00000176397.1 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr2_-_17460610 | 1.41 |
ENSMUST00000145492.1
|
Nebl
|
nebulette |
| chr13_-_67451585 | 1.39 |
ENSMUST00000057241.8
ENSMUST00000075255.5 |
Zfp874a
|
zinc finger protein 874a |
| chr12_+_10390756 | 1.39 |
ENSMUST00000020947.5
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr1_+_44147847 | 1.38 |
ENSMUST00000027214.3
|
Ercc5
|
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
| chr3_-_96452306 | 1.36 |
ENSMUST00000093126.4
ENSMUST00000098841.3 |
BC107364
|
cDNA sequence BC107364 |
| chr9_+_119341294 | 1.35 |
ENSMUST00000039784.5
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr6_-_136922169 | 1.34 |
ENSMUST00000032343.6
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr19_+_25406661 | 1.34 |
ENSMUST00000146647.1
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr4_-_42661893 | 1.29 |
ENSMUST00000108006.3
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr1_-_56969864 | 1.29 |
ENSMUST00000177424.1
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr4_+_147507600 | 1.24 |
ENSMUST00000063704.7
|
Gm13152
|
predicted gene 13152 |
| chr14_+_69347587 | 1.23 |
ENSMUST00000064831.5
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr2_+_154257854 | 1.23 |
ENSMUST00000088924.5
|
Bpifb9a
|
BPI fold containing family B, member 9A |
| chr8_-_61902669 | 1.22 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr6_-_129100903 | 1.19 |
ENSMUST00000032258.6
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr10_+_23851454 | 1.19 |
ENSMUST00000020190.7
|
Vnn3
|
vanin 3 |
| chr12_+_11438191 | 1.15 |
ENSMUST00000181268.1
|
4930511A02Rik
|
RIKEN cDNA 4930511A02 gene |
| chr4_+_105157339 | 1.15 |
ENSMUST00000064139.7
|
Ppap2b
|
phosphatidic acid phosphatase type 2B |
| chr14_-_100149764 | 1.13 |
ENSMUST00000097079.4
|
Klf12
|
Kruppel-like factor 12 |
| chr17_+_26715644 | 1.12 |
ENSMUST00000062519.7
ENSMUST00000144221.1 ENSMUST00000142539.1 ENSMUST00000151681.1 |
Crebrf
|
CREB3 regulatory factor |
| chr7_+_121083414 | 1.11 |
ENSMUST00000168311.1
ENSMUST00000171880.1 |
Otoa
|
otoancorin |
| chr3_+_41563356 | 1.10 |
ENSMUST00000163764.1
|
Phf17
|
PHD finger protein 17 |
| chr14_-_51195923 | 1.07 |
ENSMUST00000051274.1
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr15_-_37459327 | 1.06 |
ENSMUST00000119730.1
ENSMUST00000120746.1 |
Ncald
|
neurocalcin delta |
| chr11_+_58171648 | 1.05 |
ENSMUST00000020820.1
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr1_+_194619815 | 1.04 |
ENSMUST00000027952.5
|
Plxna2
|
plexin A2 |
| chr4_+_147492417 | 1.04 |
ENSMUST00000105721.2
|
Gm13152
|
predicted gene 13152 |
| chr17_+_78916473 | 1.03 |
ENSMUST00000063817.4
ENSMUST00000180077.1 |
1110001A16Rik
|
RIKEN cDNA 1110001A16 gene |
| chr12_+_88027004 | 1.02 |
ENSMUST00000178852.1
|
Gm2056
|
predicted gene 2056 |
| chr8_+_72219726 | 1.02 |
ENSMUST00000003123.8
|
Fam32a
|
family with sequence similarity 32, member A |
| chr18_-_73754457 | 1.01 |
ENSMUST00000041138.2
|
Elac1
|
elaC homolog 1 (E. coli) |
| chr8_+_88118779 | 0.99 |
ENSMUST00000121097.1
ENSMUST00000117775.1 |
Cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr16_-_48771956 | 0.97 |
ENSMUST00000170861.1
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr10_-_85127977 | 0.96 |
ENSMUST00000050813.2
|
Mterfd3
|
MTERF domain containing 3 |
| chr15_+_25773985 | 0.95 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr6_+_41538218 | 0.92 |
ENSMUST00000103291.1
|
Trbc1
|
T cell receptor beta, constant region 1 |
| chr1_-_156034800 | 0.90 |
ENSMUST00000169241.1
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr3_-_92827239 | 0.90 |
ENSMUST00000072363.4
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr17_+_70561739 | 0.90 |
ENSMUST00000097288.2
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr11_+_69838514 | 0.89 |
ENSMUST00000133967.1
ENSMUST00000094065.4 |
Tmem256
|
transmembrane protein 256 |
| chr15_+_99295087 | 0.89 |
ENSMUST00000128352.1
ENSMUST00000145482.1 |
Prpf40b
|
PRP40 pre-mRNA processing factor 40 homolog B (yeast) |
| chr6_-_113434757 | 0.87 |
ENSMUST00000113091.1
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chrX_+_73675500 | 0.86 |
ENSMUST00000171398.1
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr5_-_24902315 | 0.84 |
ENSMUST00000131486.1
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr4_-_14621669 | 0.84 |
ENSMUST00000143105.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr10_+_116966274 | 0.83 |
ENSMUST00000033651.3
|
D630029K05Rik
|
RIKEN cDNA D630029K05 gene |
| chr5_+_87925624 | 0.81 |
ENSMUST00000113271.2
|
Csn3
|
casein kappa |
| chr10_+_61171954 | 0.81 |
ENSMUST00000122261.1
ENSMUST00000121297.1 ENSMUST00000035894.5 |
Tbata
|
thymus, brain and testes associated |
| chr1_+_88306731 | 0.80 |
ENSMUST00000040210.7
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr5_-_104828957 | 0.80 |
ENSMUST00000089906.6
|
Zfp951
|
zinc finger protein 951 |
| chr6_-_108185552 | 0.79 |
ENSMUST00000167338.1
ENSMUST00000172188.1 ENSMUST00000032191.9 |
Sumf1
|
sulfatase modifying factor 1 |
| chr6_+_134640940 | 0.79 |
ENSMUST00000062755.8
|
Loh12cr1
|
loss of heterozygosity, 12, chromosomal region 1 homolog (human) |
| chr10_-_97726755 | 0.79 |
ENSMUST00000166373.1
|
C030005K15Rik
|
RIKEN cDNA C030005K15 gene |
| chrY_-_1245753 | 0.79 |
ENSMUST00000154004.1
|
Uty
|
ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome |
| chr5_-_107875035 | 0.78 |
ENSMUST00000138111.1
ENSMUST00000112642.1 |
Evi5
|
ecotropic viral integration site 5 |
| chr3_-_51408925 | 0.78 |
ENSMUST00000038108.6
|
Ndufc1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 |
| chr2_-_151668533 | 0.77 |
ENSMUST00000180195.1
ENSMUST00000096439.3 |
Rad21l
|
RAD21-like (S. pombe) |
| chrX_+_93156154 | 0.72 |
ENSMUST00000088134.3
|
1700003E24Rik
|
RIKEN cDNA 1700003E24 gene |
| chr10_+_69208546 | 0.72 |
ENSMUST00000164034.1
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr16_-_44333135 | 0.70 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr19_-_44552831 | 0.70 |
ENSMUST00000166808.1
|
Gm20538
|
predicted gene 20538 |
| chr2_+_125136692 | 0.69 |
ENSMUST00000099452.2
|
Ctxn2
|
cortexin 2 |
| chr8_+_88118747 | 0.69 |
ENSMUST00000095214.3
|
Cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr4_+_139653538 | 0.68 |
ENSMUST00000030510.7
ENSMUST00000166773.1 |
Tas1r2
|
taste receptor, type 1, member 2 |
| chrM_+_8600 | 0.66 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr12_+_116275386 | 0.66 |
ENSMUST00000090195.4
|
Gm11027
|
predicted gene 11027 |
| chr2_+_22227503 | 0.66 |
ENSMUST00000044749.7
|
Myo3a
|
myosin IIIA |
| chr15_-_35938009 | 0.61 |
ENSMUST00000156915.1
|
Cox6c
|
cytochrome c oxidase subunit VIc |
| chr4_+_47386216 | 0.59 |
ENSMUST00000107725.2
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr12_-_88274500 | 0.58 |
ENSMUST00000101168.3
|
Gm5662
|
predicted gene 5662 |
| chr16_-_44332925 | 0.58 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr9_+_85312773 | 0.58 |
ENSMUST00000113205.1
|
Gm11114
|
predicted gene 11114 |
| chr8_-_54724317 | 0.57 |
ENSMUST00000129132.2
ENSMUST00000150488.1 ENSMUST00000127511.2 |
Wdr17
|
WD repeat domain 17 |
| chr5_+_15516489 | 0.57 |
ENSMUST00000178227.1
|
Gm21847
|
predicted gene, 21847 |
| chr6_-_89940507 | 0.56 |
ENSMUST00000054202.3
|
Vmn1r45
|
vomeronasal 1 receptor 45 |
| chr4_-_86669492 | 0.54 |
ENSMUST00000149700.1
|
Plin2
|
perilipin 2 |
| chr4_-_94928789 | 0.54 |
ENSMUST00000030309.5
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr3_-_62506970 | 0.53 |
ENSMUST00000029336.4
|
Dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr3_+_41742615 | 0.53 |
ENSMUST00000146165.1
ENSMUST00000119572.1 ENSMUST00000108065.2 ENSMUST00000120167.1 ENSMUST00000026867.7 ENSMUST00000026868.7 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr2_+_24276909 | 0.52 |
ENSMUST00000028360.1
ENSMUST00000168941.1 ENSMUST00000123053.1 |
Il1f5
|
interleukin 1 family, member 5 (delta) |
| chr10_-_63927434 | 0.52 |
ENSMUST00000079279.3
|
Gm10118
|
predicted gene 10118 |
| chr7_-_103788275 | 0.51 |
ENSMUST00000183254.1
|
Olfr67
|
olfactory receptor 67 |
| chr8_+_21839926 | 0.51 |
ENSMUST00000006745.3
|
Defb2
|
defensin beta 2 |
| chr6_+_51544513 | 0.51 |
ENSMUST00000179365.1
ENSMUST00000114439.1 |
Snx10
|
sorting nexin 10 |
| chr2_-_177267036 | 0.50 |
ENSMUST00000108963.1
|
Gm14409
|
predicted gene 14409 |
| chr6_+_124024758 | 0.50 |
ENSMUST00000032238.3
|
Vmn2r26
|
vomeronasal 2, receptor 26 |
| chr1_-_72874877 | 0.49 |
ENSMUST00000027377.8
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr5_+_87666200 | 0.48 |
ENSMUST00000094641.4
|
Csn1s1
|
casein alpha s1 |
| chr8_-_21906412 | 0.47 |
ENSMUST00000051965.4
|
Defb11
|
defensin beta 11 |
| chr4_+_101986626 | 0.47 |
ENSMUST00000106914.1
|
Gm12789
|
predicted gene 12789 |
| chr9_-_107872403 | 0.47 |
ENSMUST00000183035.1
|
Rbm6
|
RNA binding motif protein 6 |
| chr2_-_129048172 | 0.47 |
ENSMUST00000145798.2
|
Gm14025
|
predicted gene 14025 |
| chr10_+_116964707 | 0.46 |
ENSMUST00000176050.1
ENSMUST00000176455.1 |
D630029K05Rik
|
RIKEN cDNA D630029K05 gene |
| chr6_+_145934113 | 0.45 |
ENSMUST00000032383.7
|
Sspn
|
sarcospan |
| chr14_+_27039001 | 0.45 |
ENSMUST00000035336.3
|
Il17rd
|
interleukin 17 receptor D |
| chr3_-_33082004 | 0.44 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr9_+_18409094 | 0.44 |
ENSMUST00000098973.2
|
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr17_-_23312313 | 0.44 |
ENSMUST00000168033.1
|
Vmn2r114
|
vomeronasal 2, receptor 114 |
| chrY_+_80135210 | 0.43 |
ENSMUST00000179811.1
|
Gm21760
|
predicted gene, 21760 |
| chr14_-_62984502 | 0.43 |
ENSMUST00000100493.2
|
Defb48
|
defensin beta 48 |
| chr16_-_35939082 | 0.42 |
ENSMUST00000081933.7
ENSMUST00000114885.1 |
Dtx3l
|
deltex 3-like (Drosophila) |
| chr17_-_32822200 | 0.42 |
ENSMUST00000179695.1
|
Zfp799
|
zinc finger protein 799 |
| chr1_-_139377094 | 0.40 |
ENSMUST00000131586.1
ENSMUST00000145244.1 |
Crb1
|
crumbs homolog 1 (Drosophila) |
| chr4_-_14621494 | 0.40 |
ENSMUST00000149633.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr15_-_35938186 | 0.40 |
ENSMUST00000014457.8
|
Cox6c
|
cytochrome c oxidase subunit VIc |
| chr10_-_78718270 | 0.40 |
ENSMUST00000005488.7
|
Casp14
|
caspase 14 |
| chr7_+_28881656 | 0.39 |
ENSMUST00000066880.4
|
Capn12
|
calpain 12 |
| chr12_+_119443410 | 0.39 |
ENSMUST00000048880.6
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr6_+_125049903 | 0.38 |
ENSMUST00000112413.1
|
Acrbp
|
proacrosin binding protein |
| chr16_-_29378667 | 0.38 |
ENSMUST00000143373.1
ENSMUST00000075806.4 |
Atp13a5
|
ATPase type 13A5 |
| chr2_-_170602017 | 0.38 |
ENSMUST00000062355.1
|
4930470P17Rik
|
RIKEN cDNA 4930470P17 gene |
| chr10_+_88379217 | 0.38 |
ENSMUST00000130301.1
ENSMUST00000020251.8 |
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr6_+_78405148 | 0.37 |
ENSMUST00000023906.2
|
Reg2
|
regenerating islet-derived 2 |
| chr12_+_88329254 | 0.36 |
ENSMUST00000091715.1
|
Gm10264
|
predicted gene 10264 |
| chr3_-_62605140 | 0.35 |
ENSMUST00000058535.5
|
Gpr149
|
G protein-coupled receptor 149 |
| chr4_+_146156824 | 0.35 |
ENSMUST00000168483.2
|
Zfp600
|
zinc finger protein 600 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Isl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 25.1 | GO:0034031 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 1.8 | 25.3 | GO:0015747 | urate transport(GO:0015747) |
| 1.5 | 4.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.8 | 9.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 4.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.8 | 2.3 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.6 | 9.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.5 | 2.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 1.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 | 1.4 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.4 | 1.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 2.6 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.4 | 14.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 1.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 1.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 1.0 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.3 | 2.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 1.5 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 | 0.9 | GO:0015881 | creatine transport(GO:0015881) |
| 0.2 | 2.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 1.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 2.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 4.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 1.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 0.6 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.2 | 1.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 1.1 | GO:1900170 | prolactin signaling pathway(GO:0038161) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 1.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 1.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.8 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 1.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 2.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.8 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.5 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 2.0 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.8 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 1.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.5 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.1 | 0.8 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 2.8 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.3 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 1.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 2.1 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 2.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.2 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 2.8 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 4.6 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 3.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 3.9 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.6 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 8.1 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 1.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.8 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 2.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.5 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 1.1 | GO:0071333 | cellular response to glucose stimulus(GO:0071333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.6 | 1.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.2 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.3 | 4.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.6 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.2 | 0.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 0.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 11.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 29.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.8 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 2.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 2.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.3 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 4.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.9 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 6.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 4.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.8 | 5.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 1.7 | 25.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.5 | 8.9 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 1.2 | 4.9 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.1 | 6.9 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.6 | 2.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.5 | 3.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.5 | 18.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 2.8 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) CoA-transferase activity(GO:0008410) palmitoyl-CoA oxidase activity(GO:0016401) C-acetyltransferase activity(GO:0016453) |
| 0.4 | 2.8 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.4 | 10.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 2.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 0.9 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.3 | 1.4 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.3 | 7.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 1.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 2.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 14.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 2.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.5 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 3.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.4 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 2.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.0 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 2.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 4.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 3.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 3.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 17.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 4.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |