Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Klf13
Z-value: 0.62
Transcription factors associated with Klf13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf13
|
ENSMUSG00000052040.9 | Kruppel-like factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf13 | mm10_v2_chr7_-_63938862_63938933 | -0.45 | 6.3e-03 | Click! |
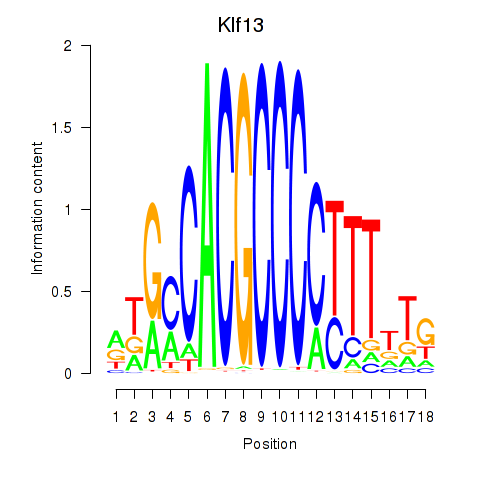
Activity profile of Klf13 motif
Sorted Z-values of Klf13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_22920222 | 4.52 |
ENSMUST00000023587.4
ENSMUST00000116625.2 |
Fetub
|
fetuin beta |
| chr12_-_104044431 | 3.60 |
ENSMUST00000043915.3
|
Serpina12
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr4_+_136143497 | 2.33 |
ENSMUST00000008016.2
|
Id3
|
inhibitor of DNA binding 3 |
| chr11_+_101665541 | 2.18 |
ENSMUST00000039388.2
|
Arl4d
|
ADP-ribosylation factor-like 4D |
| chr4_-_117133953 | 2.06 |
ENSMUST00000076859.5
|
Plk3
|
polo-like kinase 3 |
| chr9_+_57697612 | 1.89 |
ENSMUST00000034865.4
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr5_+_35757875 | 1.66 |
ENSMUST00000101280.3
ENSMUST00000054598.5 ENSMUST00000114205.1 ENSMUST00000114206.2 |
Ablim2
|
actin-binding LIM protein 2 |
| chr19_-_42129043 | 1.64 |
ENSMUST00000018965.3
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr12_-_79172609 | 1.64 |
ENSMUST00000055262.6
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr19_-_42128982 | 1.57 |
ENSMUST00000161873.1
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr12_+_108334341 | 1.57 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr7_+_25619404 | 1.52 |
ENSMUST00000077338.5
ENSMUST00000085953.3 |
Atp5sl
|
ATP5S-like |
| chr5_+_35757951 | 1.47 |
ENSMUST00000114204.1
ENSMUST00000129347.1 |
Ablim2
|
actin-binding LIM protein 2 |
| chr6_-_119417479 | 1.46 |
ENSMUST00000032272.6
|
Adipor2
|
adiponectin receptor 2 |
| chr7_-_30592941 | 1.45 |
ENSMUST00000108151.2
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr14_-_47189406 | 1.43 |
ENSMUST00000089959.6
|
Gch1
|
GTP cyclohydrolase 1 |
| chr4_-_142015056 | 1.41 |
ENSMUST00000105780.1
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr2_+_120977017 | 1.39 |
ENSMUST00000067582.7
|
Tmem62
|
transmembrane protein 62 |
| chr4_-_150008977 | 1.34 |
ENSMUST00000030830.3
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr7_+_101663705 | 1.29 |
ENSMUST00000106998.1
|
Clpb
|
ClpB caseinolytic peptidase B |
| chr6_+_137754529 | 1.26 |
ENSMUST00000087675.6
|
Dera
|
2-deoxyribose-5-phosphate aldolase homolog (C. elegans) |
| chr7_+_101663633 | 1.25 |
ENSMUST00000001884.7
|
Clpb
|
ClpB caseinolytic peptidase B |
| chr5_+_135689036 | 1.17 |
ENSMUST00000005651.6
ENSMUST00000122113.1 |
Por
|
P450 (cytochrome) oxidoreductase |
| chr11_-_119040905 | 1.17 |
ENSMUST00000026663.7
|
Cbx8
|
chromobox 8 |
| chr4_-_119422355 | 1.16 |
ENSMUST00000106316.1
ENSMUST00000030385.6 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr19_+_46761578 | 1.15 |
ENSMUST00000077666.4
ENSMUST00000099373.4 |
Cnnm2
|
cyclin M2 |
| chr11_+_67025144 | 1.11 |
ENSMUST00000079077.5
ENSMUST00000061786.5 |
Tmem220
|
transmembrane protein 220 |
| chr8_-_71486037 | 1.02 |
ENSMUST00000093450.4
|
Ano8
|
anoctamin 8 |
| chr17_-_34028044 | 1.01 |
ENSMUST00000045467.7
ENSMUST00000114303.3 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr10_-_24712133 | 1.00 |
ENSMUST00000105520.1
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr12_-_72085393 | 1.00 |
ENSMUST00000019862.2
|
L3hypdh
|
L-3-hydroxyproline dehydratase (trans-) |
| chr4_+_119422806 | 0.86 |
ENSMUST00000094819.4
|
Zmynd12
|
zinc finger, MYND domain containing 12 |
| chr16_+_4726357 | 0.86 |
ENSMUST00000154117.1
ENSMUST00000004172.8 |
Hmox2
|
heme oxygenase (decycling) 2 |
| chr9_-_31211805 | 0.86 |
ENSMUST00000072634.7
ENSMUST00000079758.7 |
Aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr10_-_24711987 | 0.85 |
ENSMUST00000135846.1
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr7_-_30563184 | 0.84 |
ENSMUST00000043898.6
|
Psenen
|
presenilin enhancer 2 homolog (C. elegans) |
| chr7_-_16614937 | 0.82 |
ENSMUST00000171937.1
ENSMUST00000075845.4 |
Grlf1
|
glucocorticoid receptor DNA binding factor 1 |
| chr1_-_36445248 | 0.82 |
ENSMUST00000125304.1
ENSMUST00000115011.1 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr11_-_120824098 | 0.79 |
ENSMUST00000055655.7
|
Fasn
|
fatty acid synthase |
| chr7_+_107595051 | 0.79 |
ENSMUST00000040056.7
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr7_-_16874845 | 0.76 |
ENSMUST00000181501.1
|
9330104G04Rik
|
RIKEN cDNA 9330104G04 gene |
| chr2_-_35200923 | 0.74 |
ENSMUST00000028238.8
ENSMUST00000113025.1 |
Rab14
|
RAB14, member RAS oncogene family |
| chr7_+_43444104 | 0.72 |
ENSMUST00000004729.3
|
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr1_+_164796723 | 0.70 |
ENSMUST00000027861.4
|
Dpt
|
dermatopontin |
| chr7_+_80246529 | 0.67 |
ENSMUST00000107381.1
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr9_-_78587968 | 0.57 |
ENSMUST00000117645.1
ENSMUST00000119213.1 ENSMUST00000052441.5 |
Slc17a5
|
solute carrier family 17 (anion/sugar transporter), member 5 |
| chr6_-_82652856 | 0.57 |
ENSMUST00000160281.1
ENSMUST00000095786.5 |
Pole4
|
polymerase (DNA-directed), epsilon 4 (p12 subunit) |
| chr17_+_56717759 | 0.56 |
ENSMUST00000002452.6
|
Ndufa11
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 11 |
| chr7_+_3694512 | 0.55 |
ENSMUST00000108627.3
|
Tsen34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae) |
| chr4_-_126736236 | 0.55 |
ENSMUST00000048194.7
|
Tfap2e
|
transcription factor AP-2, epsilon |
| chr11_+_82388900 | 0.55 |
ENSMUST00000054245.4
ENSMUST00000092852.2 |
Tmem132e
|
transmembrane protein 132E |
| chr13_-_49215978 | 0.55 |
ENSMUST00000048946.6
|
1110007C09Rik
|
RIKEN cDNA 1110007C09 gene |
| chr17_-_56757516 | 0.55 |
ENSMUST00000044752.5
|
Nrtn
|
neurturin |
| chr14_+_122107038 | 0.52 |
ENSMUST00000026624.4
|
Tm9sf2
|
transmembrane 9 superfamily member 2 |
| chr18_+_67343564 | 0.52 |
ENSMUST00000025404.8
|
Cidea
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| chr17_-_87282793 | 0.51 |
ENSMUST00000146560.2
|
4833418N02Rik
|
RIKEN cDNA 4833418N02 gene |
| chr17_-_56626872 | 0.51 |
ENSMUST00000047226.8
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr6_+_83795205 | 0.51 |
ENSMUST00000113850.1
|
Nagk
|
N-acetylglucosamine kinase |
| chr17_-_87282771 | 0.48 |
ENSMUST00000161759.1
|
4833418N02Rik
|
RIKEN cDNA 4833418N02 gene |
| chr2_+_167538192 | 0.48 |
ENSMUST00000052631.7
|
Snai1
|
snail homolog 1 (Drosophila) |
| chr7_+_80246375 | 0.46 |
ENSMUST00000058266.6
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr6_+_83794974 | 0.46 |
ENSMUST00000037376.7
|
Nagk
|
N-acetylglucosamine kinase |
| chr17_+_87282880 | 0.45 |
ENSMUST00000041110.5
ENSMUST00000125875.1 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr17_-_35909626 | 0.43 |
ENSMUST00000141132.1
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr7_+_30563298 | 0.43 |
ENSMUST00000043273.9
ENSMUST00000163654.1 ENSMUST00000166510.1 ENSMUST00000163482.1 ENSMUST00000166257.1 ENSMUST00000163464.1 ENSMUST00000171850.1 ENSMUST00000168931.1 |
U2af1l4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr14_+_122107119 | 0.42 |
ENSMUST00000171318.1
|
Tm9sf2
|
transmembrane 9 superfamily member 2 |
| chr7_+_19181159 | 0.37 |
ENSMUST00000120595.1
ENSMUST00000048502.8 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr6_+_83795022 | 0.37 |
ENSMUST00000113851.1
|
Nagk
|
N-acetylglucosamine kinase |
| chr14_-_55591077 | 0.36 |
ENSMUST00000161807.1
ENSMUST00000111378.3 ENSMUST00000159687.1 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr7_-_144582392 | 0.34 |
ENSMUST00000033394.7
|
Fadd
|
Fas (TNFRSF6)-associated via death domain |
| chr9_-_75409951 | 0.32 |
ENSMUST00000049355.10
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr5_-_115158169 | 0.32 |
ENSMUST00000053271.5
ENSMUST00000112121.1 |
Mlec
|
malectin |
| chr3_-_116711820 | 0.31 |
ENSMUST00000153108.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr12_+_72085847 | 0.28 |
ENSMUST00000117449.1
ENSMUST00000057257.8 |
Jkamp
|
JNK1/MAPK8-associated membrane protein |
| chrX_-_137120128 | 0.28 |
ENSMUST00000113066.2
|
Esx1
|
extraembryonic, spermatogenesis, homeobox 1 |
| chr14_+_54426902 | 0.28 |
ENSMUST00000010550.7
|
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr9_+_75410145 | 0.27 |
ENSMUST00000180533.1
ENSMUST00000180574.1 |
4933433G15Rik
|
RIKEN cDNA 4933433G15 gene |
| chr14_-_76010863 | 0.27 |
ENSMUST00000088922.4
|
Gtf2f2
|
general transcription factor IIF, polypeptide 2 |
| chr7_-_27178835 | 0.27 |
ENSMUST00000093040.6
|
Rab4b
|
RAB4B, member RAS oncogene family |
| chr2_-_26445175 | 0.25 |
ENSMUST00000114082.2
ENSMUST00000091252.4 |
Sec16a
|
SEC16 homolog A (S. cerevisiae) |
| chr10_-_40257648 | 0.21 |
ENSMUST00000019982.7
|
Gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr2_-_73892619 | 0.20 |
ENSMUST00000112007.1
ENSMUST00000112016.2 |
Atf2
|
activating transcription factor 2 |
| chr15_-_64922290 | 0.17 |
ENSMUST00000023007.5
|
Adcy8
|
adenylate cyclase 8 |
| chr2_-_137116624 | 0.16 |
ENSMUST00000028735.7
|
Jag1
|
jagged 1 |
| chr10_-_81167896 | 0.16 |
ENSMUST00000005064.7
|
Pias4
|
protein inhibitor of activated STAT 4 |
| chr8_-_71558871 | 0.15 |
ENSMUST00000052072.6
|
Tmem221
|
transmembrane protein 221 |
| chr8_+_84990585 | 0.15 |
ENSMUST00000064495.6
|
Hook2
|
hook homolog 2 (Drosophila) |
| chr18_+_37955544 | 0.15 |
ENSMUST00000070709.2
ENSMUST00000177058.1 ENSMUST00000169360.2 ENSMUST00000163591.2 ENSMUST00000091932.5 |
Rell2
|
RELT-like 2 |
| chr2_-_73892588 | 0.13 |
ENSMUST00000154456.1
ENSMUST00000090802.4 ENSMUST00000055833.5 |
Atf2
|
activating transcription factor 2 |
| chr7_+_18925863 | 0.12 |
ENSMUST00000172835.1
ENSMUST00000032571.8 |
Nova2
|
neuro-oncological ventral antigen 2 |
| chr7_+_16875302 | 0.12 |
ENSMUST00000108493.1
|
Dact3
|
dapper homolog 3, antagonist of beta-catenin (xenopus) |
| chr5_+_72914554 | 0.11 |
ENSMUST00000143829.1
|
Slain2
|
SLAIN motif family, member 2 |
| chr5_-_138263942 | 0.10 |
ENSMUST00000048421.7
ENSMUST00000164203.1 |
BC037034
BC037034
|
cDNA sequence BC037034 cDNA sequence BC037034 |
| chr19_-_5610038 | 0.09 |
ENSMUST00000113641.2
|
Kat5
|
K(lysine) acetyltransferase 5 |
| chr10_+_126978690 | 0.08 |
ENSMUST00000105256.2
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr5_-_137502402 | 0.07 |
ENSMUST00000111035.1
ENSMUST00000031728.4 |
Pop7
|
processing of precursor 7, ribonuclease P family, (S. cerevisiae) |
| chr1_+_128244122 | 0.07 |
ENSMUST00000027592.3
|
Ubxn4
|
UBX domain protein 4 |
| chr18_-_37954958 | 0.07 |
ENSMUST00000043498.7
|
Hdac3
|
histone deacetylase 3 |
| chr6_+_71293765 | 0.04 |
ENSMUST00000114185.2
|
Gm1070
|
predicted gene 1070 |
| chr2_-_73892530 | 0.04 |
ENSMUST00000136958.1
ENSMUST00000112010.2 ENSMUST00000128531.1 ENSMUST00000112017.1 |
Atf2
|
activating transcription factor 2 |
| chrX_-_8252334 | 0.04 |
ENSMUST00000115595.1
ENSMUST00000033513.3 |
Ftsj1
|
FtsJ homolog 1 (E. coli) |
| chr3_-_90509450 | 0.03 |
ENSMUST00000107343.1
ENSMUST00000001043.7 ENSMUST00000107344.1 ENSMUST00000076639.4 ENSMUST00000107346.1 ENSMUST00000146740.1 ENSMUST00000107342.1 ENSMUST00000049937.6 |
Chtop
|
chromatin target of PRMT1 |
| chr5_+_72914264 | 0.02 |
ENSMUST00000144843.1
|
Slain2
|
SLAIN motif family, member 2 |
| chr19_-_4811508 | 0.02 |
ENSMUST00000179909.1
ENSMUST00000172000.2 ENSMUST00000180008.1 ENSMUST00000113793.3 ENSMUST00000006625.7 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr3_-_142395661 | 0.02 |
ENSMUST00000029941.9
ENSMUST00000168967.2 ENSMUST00000090134.5 ENSMUST00000058626.8 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr7_+_5020376 | 0.02 |
ENSMUST00000076251.4
|
Zfp865
|
zinc finger protein 865 |
| chr5_-_38220400 | 0.01 |
ENSMUST00000114113.1
ENSMUST00000094833.3 |
Zbtb49
|
zinc finger and BTB domain containing 49 |
| chr2_+_32288244 | 0.00 |
ENSMUST00000113377.1
ENSMUST00000100194.2 |
Golga2
|
golgi autoantigen, golgin subfamily a, 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0018894 | coumarin metabolic process(GO:0009804) dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.4 | 1.4 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.3 | 1.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.3 | 1.2 | GO:0090346 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 3.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 2.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 0.9 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.2 | 1.9 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.2 | 1.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 1.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.1 | 1.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 4.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.8 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.4 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 2.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 1.2 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 2.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 1.2 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 2.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.6 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.7 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.4 | 1.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.4 | 1.5 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.3 | 1.0 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.3 | 1.9 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 4.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.2 | GO:0047726 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 1.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.9 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.9 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.2 | 1.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 0.8 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 1.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 1.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 2.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 2.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |