Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Klf7
Z-value: 0.73
Transcription factors associated with Klf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf7
|
ENSMUSG00000025959.7 | Kruppel-like factor 7 (ubiquitous) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf7 | mm10_v2_chr1_-_64121456_64121491 | 0.28 | 9.2e-02 | Click! |
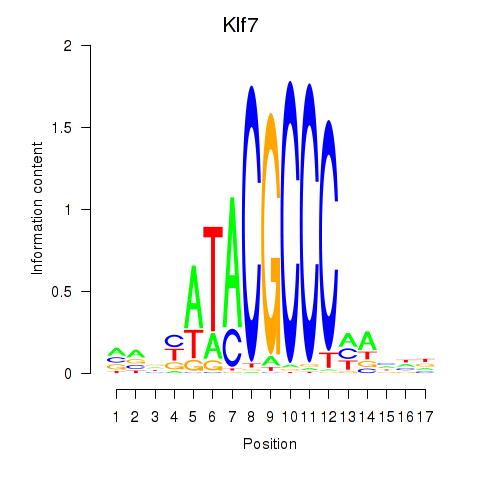
Activity profile of Klf7 motif
Sorted Z-values of Klf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102107822 | 7.24 |
ENSMUST00000177304.1
ENSMUST00000017455.8 |
Pyy
|
peptide YY |
| chrX_+_136270253 | 4.17 |
ENSMUST00000178632.1
ENSMUST00000053540.4 |
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr8_+_84901928 | 3.02 |
ENSMUST00000067060.7
|
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr2_+_119047116 | 2.81 |
ENSMUST00000152380.1
ENSMUST00000099542.2 |
Casc5
|
cancer susceptibility candidate 5 |
| chr15_+_99074968 | 2.52 |
ENSMUST00000039665.6
|
Troap
|
trophinin associated protein |
| chr5_-_137741601 | 2.36 |
ENSMUST00000119498.1
ENSMUST00000061789.7 |
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr6_-_88875035 | 2.35 |
ENSMUST00000145944.1
|
Podxl2
|
podocalyxin-like 2 |
| chr5_+_33658123 | 2.18 |
ENSMUST00000074849.6
ENSMUST00000079534.4 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr2_+_119047129 | 2.18 |
ENSMUST00000153300.1
ENSMUST00000028799.5 |
Casc5
|
cancer susceptibility candidate 5 |
| chr5_+_137761680 | 1.93 |
ENSMUST00000110983.2
ENSMUST00000031738.4 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr11_-_61494173 | 1.89 |
ENSMUST00000101085.2
ENSMUST00000079080.6 ENSMUST00000108714.1 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr7_-_127993831 | 1.72 |
ENSMUST00000033056.3
|
Pycard
|
PYD and CARD domain containing |
| chr13_+_21722057 | 1.68 |
ENSMUST00000110476.3
|
Hist1h2bm
|
histone cluster 1, H2bm |
| chr6_-_88874597 | 1.65 |
ENSMUST00000061262.4
ENSMUST00000140455.1 ENSMUST00000145780.1 |
Podxl2
|
podocalyxin-like 2 |
| chr3_+_51224447 | 1.63 |
ENSMUST00000023849.8
ENSMUST00000167780.1 |
Ccrn4l
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
| chr7_-_132776855 | 1.52 |
ENSMUST00000106168.1
|
Fam53b
|
family with sequence similarity 53, member B |
| chrX_-_74428871 | 1.51 |
ENSMUST00000143521.1
|
G6pdx
|
glucose-6-phosphate dehydrogenase X-linked |
| chr19_-_4059295 | 1.48 |
ENSMUST00000076451.6
|
BC021614
|
cDNA sequence BC021614 |
| chr7_-_139978748 | 1.48 |
ENSMUST00000097970.2
|
6430531B16Rik
|
RIKEN cDNA 6430531B16 gene |
| chr5_-_136135989 | 1.43 |
ENSMUST00000150406.1
ENSMUST00000006301.4 |
Lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr8_+_119992438 | 1.40 |
ENSMUST00000132583.1
ENSMUST00000034282.9 |
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr12_-_4592927 | 1.37 |
ENSMUST00000170816.1
|
Gm3625
|
predicted gene 3625 |
| chr5_+_44100442 | 1.35 |
ENSMUST00000072800.4
|
Gm16401
|
predicted gene 16401 |
| chr7_-_74013676 | 1.32 |
ENSMUST00000026896.4
|
St8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr8_+_70152754 | 1.32 |
ENSMUST00000072500.6
ENSMUST00000164040.1 ENSMUST00000110146.2 ENSMUST00000110143.1 ENSMUST00000110141.2 ENSMUST00000110140.1 |
2310045N01Rik
Mef2b
|
RIKEN cDNA 2310045N01 gene myocyte enhancer factor 2B |
| chr7_-_139978709 | 1.23 |
ENSMUST00000121412.1
|
6430531B16Rik
|
RIKEN cDNA 6430531B16 gene |
| chr6_+_120093348 | 1.17 |
ENSMUST00000112711.2
|
Ninj2
|
ninjurin 2 |
| chr15_+_92344359 | 1.14 |
ENSMUST00000181901.1
|
Gm26760
|
predicted gene, 26760 |
| chr9_-_32344237 | 1.14 |
ENSMUST00000034533.5
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr4_+_107830958 | 1.08 |
ENSMUST00000106731.2
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr4_+_63558748 | 1.03 |
ENSMUST00000077709.4
|
6330416G13Rik
|
RIKEN cDNA 6330416G13 gene |
| chr1_+_63176818 | 1.02 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr5_+_140607334 | 0.85 |
ENSMUST00000031555.1
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_+_58658181 | 0.84 |
ENSMUST00000168747.1
|
Atp10a
|
ATPase, class V, type 10A |
| chr17_-_46202576 | 0.84 |
ENSMUST00000024749.7
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr11_+_68432112 | 0.82 |
ENSMUST00000021283.7
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5, p101 |
| chr14_+_30479565 | 0.77 |
ENSMUST00000022535.7
|
Dcp1a
|
DCP1 decapping enzyme homolog A (S. cerevisiae) |
| chr7_-_19404082 | 0.72 |
ENSMUST00000108458.3
|
Klc3
|
kinesin light chain 3 |
| chr14_+_79451791 | 0.70 |
ENSMUST00000100359.1
|
Zbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr10_+_127677064 | 0.70 |
ENSMUST00000118612.1
ENSMUST00000048099.4 |
Tmem194
|
transmembrane protein 194 |
| chr15_-_98953541 | 0.67 |
ENSMUST00000097014.5
|
Tuba1a
|
tubulin, alpha 1A |
| chr5_+_136136137 | 0.63 |
ENSMUST00000136634.1
ENSMUST00000041100.3 |
Alkbh4
|
alkB, alkylation repair homolog 4 (E. coli) |
| chr18_+_88971790 | 0.62 |
ENSMUST00000023828.7
|
Rttn
|
rotatin |
| chr9_-_45936049 | 0.61 |
ENSMUST00000034590.2
|
Tagln
|
transgelin |
| chr5_+_136136202 | 0.58 |
ENSMUST00000143229.1
|
Alkbh4
|
alkB, alkylation repair homolog 4 (E. coli) |
| chr8_+_105860634 | 0.57 |
ENSMUST00000008594.7
|
Nutf2
|
nuclear transport factor 2 |
| chr7_-_126976092 | 0.54 |
ENSMUST00000181859.1
|
D830044I16Rik
|
RIKEN cDNA D830044I16 gene |
| chr8_-_3621270 | 0.51 |
ENSMUST00000159548.1
ENSMUST00000019614.6 |
Xab2
|
XPA binding protein 2 |
| chr17_+_46202740 | 0.50 |
ENSMUST00000087031.5
|
Xpo5
|
exportin 5 |
| chr2_-_151973387 | 0.49 |
ENSMUST00000109863.1
|
Fam110a
|
family with sequence similarity 110, member A |
| chr14_+_50807915 | 0.49 |
ENSMUST00000036126.5
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr5_+_115631902 | 0.47 |
ENSMUST00000031492.8
|
Rab35
|
RAB35, member RAS oncogene family |
| chr3_+_28263205 | 0.47 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr17_+_35916541 | 0.46 |
ENSMUST00000087211.2
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr11_-_40733373 | 0.43 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr12_-_79296266 | 0.43 |
ENSMUST00000021547.6
|
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr7_+_28350652 | 0.42 |
ENSMUST00000082134.4
|
Rps16
|
ribosomal protein S16 |
| chr11_-_78176619 | 0.42 |
ENSMUST00000148154.2
ENSMUST00000017549.6 |
Nek8
|
NIMA (never in mitosis gene a)-related expressed kinase 8 |
| chr17_+_35916977 | 0.42 |
ENSMUST00000151664.1
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr19_+_8723478 | 0.40 |
ENSMUST00000180819.1
ENSMUST00000181422.1 |
Snhg1
|
small nucleolar RNA host gene (non-protein coding) 1 |
| chr7_-_68749170 | 0.40 |
ENSMUST00000118110.1
ENSMUST00000048068.7 |
Arrdc4
|
arrestin domain containing 4 |
| chr9_+_55541148 | 0.38 |
ENSMUST00000034869.4
|
Isl2
|
insulin related protein 2 (islet 2) |
| chr7_+_28071230 | 0.37 |
ENSMUST00000138392.1
ENSMUST00000076648.7 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr1_+_171329015 | 0.37 |
ENSMUST00000111300.1
|
Dedd
|
death effector domain-containing |
| chr11_-_120991039 | 0.34 |
ENSMUST00000070575.7
|
Csnk1d
|
casein kinase 1, delta |
| chr12_-_11436607 | 0.31 |
ENSMUST00000072299.5
|
Vsnl1
|
visinin-like 1 |
| chr3_-_103791075 | 0.26 |
ENSMUST00000106845.2
ENSMUST00000029438.8 ENSMUST00000121324.1 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr11_+_80383279 | 0.25 |
ENSMUST00000165565.1
ENSMUST00000017567.7 |
Zfp207
|
zinc finger protein 207 |
| chr8_-_126945841 | 0.24 |
ENSMUST00000179857.1
|
Tomm20
|
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr16_+_36694024 | 0.23 |
ENSMUST00000119464.1
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr16_+_36693972 | 0.22 |
ENSMUST00000023617.6
ENSMUST00000089618.3 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr5_-_124187150 | 0.22 |
ENSMUST00000161938.1
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr9_-_36767595 | 0.21 |
ENSMUST00000120381.2
|
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr2_-_156312470 | 0.21 |
ENSMUST00000079125.6
|
Scand1
|
SCAN domain-containing 1 |
| chr9_-_71485893 | 0.17 |
ENSMUST00000034720.5
|
Polr2m
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr6_+_125039760 | 0.17 |
ENSMUST00000140131.1
ENSMUST00000032480.7 |
Ing4
|
inhibitor of growth family, member 4 |
| chr8_+_71367210 | 0.16 |
ENSMUST00000019169.7
|
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr15_-_102722120 | 0.15 |
ENSMUST00000171838.1
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr17_-_84466186 | 0.13 |
ENSMUST00000047524.8
|
Thada
|
thyroid adenoma associated |
| chr8_-_84966008 | 0.12 |
ENSMUST00000109738.3
ENSMUST00000065049.8 ENSMUST00000128972.1 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr2_-_32775378 | 0.10 |
ENSMUST00000091059.5
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr15_-_102722150 | 0.10 |
ENSMUST00000023818.3
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr11_+_70166696 | 0.08 |
ENSMUST00000178567.1
|
Clec10a
|
C-type lectin domain family 10, member A |
| chr11_-_70654624 | 0.08 |
ENSMUST00000018437.2
|
Pfn1
|
profilin 1 |
| chr17_+_24488773 | 0.06 |
ENSMUST00000024958.7
|
Caskin1
|
CASK interacting protein 1 |
| chr15_-_50889043 | 0.06 |
ENSMUST00000183997.1
ENSMUST00000183757.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr9_+_98955525 | 0.05 |
ENSMUST00000051312.2
|
Foxl2
|
forkhead box L2 |
| chr13_-_14613017 | 0.05 |
ENSMUST00000015816.3
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr15_+_30172570 | 0.04 |
ENSMUST00000081728.5
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr12_+_49383007 | 0.02 |
ENSMUST00000021333.3
|
Foxg1
|
forkhead box G1 |
| chr2_-_32775625 | 0.01 |
ENSMUST00000161958.1
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr19_+_5637475 | 0.01 |
ENSMUST00000025867.5
|
Rela
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian) |
| chr5_+_45520221 | 0.01 |
ENSMUST00000156481.1
ENSMUST00000119579.1 ENSMUST00000118833.1 |
Med28
|
mediator of RNA polymerase II transcription, subunit 28 homolog (yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.6 | 1.7 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.5 | 1.5 | GO:1904879 | pentose-phosphate shunt, oxidative branch(GO:0009051) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 7.2 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.4 | 1.2 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.2 | 3.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 1.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.6 | GO:1904046 | protein localization to nuclear pore(GO:0090204) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.2 | 1.3 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 0.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 1.1 | GO:1990573 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 4.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 2.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.5 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 1.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 1.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.8 | GO:0006290 | pyrimidine dimer repair(GO:0006290) cellular response to UV-C(GO:0071494) |
| 0.1 | 0.5 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.6 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.0 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 1.5 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 1.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 1.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.7 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 1.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.5 | GO:0042272 | nucleocytoplasmic shuttling complex(GO:0031074) nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.2 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.1 | 1.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0044233 | mitochondrial outer membrane translocase complex(GO:0005742) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 1.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.3 | 7.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.2 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 3.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 6.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 7.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 3.9 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 1.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |