Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Mef2c
Z-value: 1.16
Transcription factors associated with Mef2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2c
|
ENSMUSG00000005583.10 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2c | mm10_v2_chr13_+_83573577_83573607 | 0.41 | 1.4e-02 | Click! |
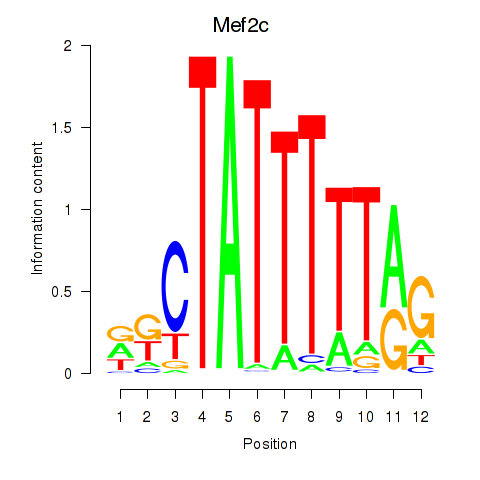
Activity profile of Mef2c motif
Sorted Z-values of Mef2c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_98383811 | 7.73 |
ENSMUST00000008021.2
|
Tcap
|
titin-cap |
| chr5_+_122100951 | 6.70 |
ENSMUST00000014080.6
ENSMUST00000111750.1 ENSMUST00000139213.1 ENSMUST00000111751.1 ENSMUST00000155612.1 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr1_-_66935333 | 6.32 |
ENSMUST00000120415.1
ENSMUST00000119429.1 |
Myl1
|
myosin, light polypeptide 1 |
| chr3_-_123034943 | 5.31 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr7_+_142498832 | 5.21 |
ENSMUST00000078497.8
ENSMUST00000105953.3 ENSMUST00000179658.1 ENSMUST00000105954.3 ENSMUST00000105952.3 ENSMUST00000105955.1 ENSMUST00000074187.6 ENSMUST00000180152.1 ENSMUST00000105950.4 ENSMUST00000105957.3 ENSMUST00000169299.2 ENSMUST00000105958.3 ENSMUST00000105949.1 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr7_+_142441808 | 4.99 |
ENSMUST00000105971.1
ENSMUST00000145287.1 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr15_-_77022632 | 4.64 |
ENSMUST00000019037.8
ENSMUST00000169226.1 |
Mb
|
myoglobin |
| chr11_+_67798269 | 4.14 |
ENSMUST00000168612.1
ENSMUST00000040574.4 |
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr14_-_34588654 | 4.14 |
ENSMUST00000022328.6
ENSMUST00000064098.6 ENSMUST00000022327.5 ENSMUST00000022330.7 |
Ldb3
|
LIM domain binding 3 |
| chr9_+_110763646 | 4.12 |
ENSMUST00000079784.7
|
Myl3
|
myosin, light polypeptide 3 |
| chr15_-_78206391 | 4.07 |
ENSMUST00000120592.1
|
Pvalb
|
parvalbumin |
| chr1_+_40805578 | 3.92 |
ENSMUST00000114765.2
|
Tmem182
|
transmembrane protein 182 |
| chr1_-_172219715 | 3.70 |
ENSMUST00000170700.1
ENSMUST00000003554.4 |
Casq1
|
calsequestrin 1 |
| chr8_+_15057646 | 3.40 |
ENSMUST00000033842.3
|
Myom2
|
myomesin 2 |
| chr7_+_142442330 | 3.29 |
ENSMUST00000149529.1
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr11_-_94976327 | 3.22 |
ENSMUST00000103162.1
ENSMUST00000166320.1 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr4_+_130308595 | 3.05 |
ENSMUST00000070532.7
|
Fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr5_-_112896350 | 2.86 |
ENSMUST00000086617.4
|
Myo18b
|
myosin XVIIIb |
| chr14_-_34588607 | 2.82 |
ENSMUST00000090040.4
|
Ldb3
|
LIM domain binding 3 |
| chr14_-_20664562 | 2.76 |
ENSMUST00000119483.1
|
Synpo2l
|
synaptopodin 2-like |
| chr7_+_102101736 | 2.71 |
ENSMUST00000033300.2
|
Art1
|
ADP-ribosyltransferase 1 |
| chrX_-_95478107 | 2.67 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr10_+_33083476 | 2.50 |
ENSMUST00000095762.4
|
Trdn
|
triadin |
| chr7_+_128003911 | 2.34 |
ENSMUST00000106248.1
|
Trim72
|
tripartite motif-containing 72 |
| chr3_+_102086471 | 2.22 |
ENSMUST00000165540.2
ENSMUST00000164123.1 |
Casq2
|
calsequestrin 2 |
| chr14_-_20668269 | 2.20 |
ENSMUST00000057090.5
ENSMUST00000117386.1 |
Synpo2l
|
synaptopodin 2-like |
| chr7_-_128206346 | 2.19 |
ENSMUST00000033049.7
|
Cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr14_-_32685246 | 2.18 |
ENSMUST00000096038.3
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene |
| chr14_-_54966570 | 2.17 |
ENSMUST00000124930.1
ENSMUST00000134256.1 ENSMUST00000081857.6 ENSMUST00000145322.1 |
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr9_+_121777607 | 2.14 |
ENSMUST00000098272.2
|
Klhl40
|
kelch-like 40 |
| chr1_+_135799402 | 2.10 |
ENSMUST00000152208.1
ENSMUST00000152075.1 ENSMUST00000154463.1 ENSMUST00000139986.1 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr3_+_102086415 | 2.07 |
ENSMUST00000029454.5
|
Casq2
|
calsequestrin 2 |
| chr11_+_77462325 | 2.05 |
ENSMUST00000102493.1
|
Coro6
|
coronin 6 |
| chr3_+_103074009 | 2.04 |
ENSMUST00000090715.6
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr14_-_21714570 | 1.91 |
ENSMUST00000073870.5
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr1_+_135799833 | 1.90 |
ENSMUST00000148201.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr11_+_67238017 | 1.86 |
ENSMUST00000018632.4
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr13_-_113663670 | 1.80 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr2_-_76982455 | 1.68 |
ENSMUST00000011934.5
ENSMUST00000099981.2 ENSMUST00000099980.3 ENSMUST00000111882.2 ENSMUST00000140091.1 |
Ttn
|
titin |
| chr6_+_30541582 | 1.66 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr2_+_69670100 | 1.65 |
ENSMUST00000100050.3
|
Klhl41
|
kelch-like 41 |
| chr3_+_103074035 | 1.65 |
ENSMUST00000176440.1
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr4_-_73950834 | 1.64 |
ENSMUST00000095023.1
ENSMUST00000030101.3 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr2_+_32628390 | 1.54 |
ENSMUST00000156578.1
|
Ak1
|
adenylate kinase 1 |
| chr17_+_71019548 | 1.44 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr11_+_70657196 | 1.42 |
ENSMUST00000157027.1
ENSMUST00000072841.5 ENSMUST00000108548.1 ENSMUST00000126241.1 |
Eno3
|
enolase 3, beta muscle |
| chr4_+_156215920 | 1.40 |
ENSMUST00000105572.1
|
2310042D19Rik
|
RIKEN cDNA 2310042D19 gene |
| chr11_-_107716517 | 1.39 |
ENSMUST00000021065.5
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr11_+_70657687 | 1.37 |
ENSMUST00000134087.1
ENSMUST00000170716.1 |
Eno3
|
enolase 3, beta muscle |
| chr2_-_104028287 | 1.33 |
ENSMUST00000056170.3
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr17_+_71019503 | 1.32 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr1_-_44019929 | 1.31 |
ENSMUST00000061421.3
|
Mettl21c
|
methyltransferase like 21C |
| chr6_+_112273758 | 1.28 |
ENSMUST00000032376.5
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr1_+_187608755 | 1.25 |
ENSMUST00000127489.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr5_+_66745835 | 1.23 |
ENSMUST00000101164.4
ENSMUST00000118242.1 ENSMUST00000119854.1 ENSMUST00000117601.1 |
Limch1
|
LIM and calponin homology domains 1 |
| chr15_+_25940846 | 1.22 |
ENSMUST00000110438.1
|
Fam134b
|
family with sequence similarity 134, member B |
| chr7_-_4514558 | 1.18 |
ENSMUST00000163538.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr19_+_6384395 | 1.18 |
ENSMUST00000035269.8
ENSMUST00000113483.1 |
Pygm
|
muscle glycogen phosphorylase |
| chr14_+_26894557 | 1.14 |
ENSMUST00000090337.4
ENSMUST00000165929.2 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr2_-_67433181 | 1.10 |
ENSMUST00000180773.1
|
Gm26727
|
predicted gene, 26727 |
| chr10_+_127501672 | 1.08 |
ENSMUST00000160019.1
ENSMUST00000160610.1 |
Stac3
|
SH3 and cysteine rich domain 3 |
| chr4_+_135759705 | 1.05 |
ENSMUST00000105854.1
|
Myom3
|
myomesin family, member 3 |
| chr5_+_122101146 | 1.03 |
ENSMUST00000147178.1
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr4_-_130308674 | 1.01 |
ENSMUST00000097865.1
|
Gm10570
|
predicted gene 10570 |
| chr7_+_45405960 | 1.00 |
ENSMUST00000107774.1
|
Kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr7_-_4514609 | 0.96 |
ENSMUST00000166959.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr9_-_120023558 | 0.96 |
ENSMUST00000111635.2
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr4_+_137862270 | 0.95 |
ENSMUST00000130407.1
|
Ece1
|
endothelin converting enzyme 1 |
| chr5_+_92386879 | 0.95 |
ENSMUST00000128246.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chrX_+_164438039 | 0.94 |
ENSMUST00000033755.5
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr4_+_49059256 | 0.93 |
ENSMUST00000076670.2
|
E130309F12Rik
|
RIKEN cDNA E130309F12 gene |
| chr5_-_62765618 | 0.92 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr10_+_127501707 | 0.90 |
ENSMUST00000035839.2
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr3_+_32817520 | 0.90 |
ENSMUST00000072312.5
ENSMUST00000108228.1 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr11_+_75656103 | 0.90 |
ENSMUST00000136935.1
|
Myo1c
|
myosin IC |
| chr2_+_91118136 | 0.90 |
ENSMUST00000169776.1
ENSMUST00000111430.3 ENSMUST00000137942.1 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr5_-_122002340 | 0.87 |
ENSMUST00000134326.1
|
Cux2
|
cut-like homeobox 2 |
| chr15_-_91191733 | 0.87 |
ENSMUST00000069511.6
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr8_+_45658273 | 0.87 |
ENSMUST00000153798.1
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_+_130865835 | 0.85 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr12_+_88953399 | 0.83 |
ENSMUST00000057634.7
|
Nrxn3
|
neurexin III |
| chr13_+_83573577 | 0.81 |
ENSMUST00000185052.1
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_+_72054598 | 0.76 |
ENSMUST00000028525.5
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_67445994 | 0.72 |
ENSMUST00000112347.1
ENSMUST00000028410.3 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr13_+_37826904 | 0.70 |
ENSMUST00000149745.1
|
Rreb1
|
ras responsive element binding protein 1 |
| chr7_+_122671378 | 0.70 |
ENSMUST00000182563.1
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr2_-_77170592 | 0.67 |
ENSMUST00000164114.2
ENSMUST00000049544.7 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr14_+_101840501 | 0.66 |
ENSMUST00000159026.1
|
Lmo7
|
LIM domain only 7 |
| chrX_+_73774397 | 0.65 |
ENSMUST00000002081.5
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr15_-_58324161 | 0.64 |
ENSMUST00000022985.1
|
Klhl38
|
kelch-like 38 |
| chr1_+_187609028 | 0.64 |
ENSMUST00000110939.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr9_+_77636494 | 0.63 |
ENSMUST00000057781.7
|
Klhl31
|
kelch-like 31 |
| chr10_+_94550852 | 0.63 |
ENSMUST00000148910.1
ENSMUST00000117460.1 |
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr5_-_51553896 | 0.61 |
ENSMUST00000132734.1
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr14_+_66344296 | 0.60 |
ENSMUST00000152093.1
ENSMUST00000074523.6 |
Stmn4
|
stathmin-like 4 |
| chr9_-_79977782 | 0.58 |
ENSMUST00000093811.4
|
Filip1
|
filamin A interacting protein 1 |
| chrX_+_106583184 | 0.58 |
ENSMUST00000101296.2
ENSMUST00000101297.3 |
Gm5127
|
predicted gene 5127 |
| chr8_+_31089471 | 0.57 |
ENSMUST00000036631.7
ENSMUST00000170204.1 |
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr6_-_142702259 | 0.56 |
ENSMUST00000073173.5
ENSMUST00000111771.1 ENSMUST00000087527.4 ENSMUST00000100827.2 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr19_+_4003334 | 0.55 |
ENSMUST00000025806.3
|
Doc2g
|
double C2, gamma |
| chr5_-_88527841 | 0.55 |
ENSMUST00000087033.3
|
Igj
|
immunoglobulin joining chain |
| chr6_+_42286676 | 0.54 |
ENSMUST00000031894.6
|
Clcn1
|
chloride channel 1 |
| chr1_+_135836380 | 0.53 |
ENSMUST00000178204.1
|
Tnnt2
|
troponin T2, cardiac |
| chr7_+_130865756 | 0.52 |
ENSMUST00000120441.1
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr9_-_65391652 | 0.52 |
ENSMUST00000068307.3
|
Kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr5_+_107497762 | 0.52 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr6_-_128526703 | 0.50 |
ENSMUST00000143664.1
ENSMUST00000112132.1 ENSMUST00000032510.7 |
Pzp
|
pregnancy zone protein |
| chrX_+_159303266 | 0.50 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr5_-_24540439 | 0.50 |
ENSMUST00000048302.6
ENSMUST00000119657.1 |
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr17_-_14694223 | 0.49 |
ENSMUST00000170872.1
|
Thbs2
|
thrombospondin 2 |
| chr7_-_4514368 | 0.49 |
ENSMUST00000166161.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr1_-_172329261 | 0.49 |
ENSMUST00000062387.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr17_-_67950908 | 0.49 |
ENSMUST00000164647.1
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr7_-_81566939 | 0.48 |
ENSMUST00000042318.5
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr10_-_69212996 | 0.48 |
ENSMUST00000170048.1
|
A930033H14Rik
|
RIKEN cDNA A930033H14 gene |
| chrX_+_56779699 | 0.48 |
ENSMUST00000114772.2
ENSMUST00000114768.3 ENSMUST00000155882.1 |
Fhl1
|
four and a half LIM domains 1 |
| chr5_-_129879038 | 0.45 |
ENSMUST00000026617.6
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr7_+_112742025 | 0.44 |
ENSMUST00000164363.1
|
Tead1
|
TEA domain family member 1 |
| chr6_+_5390387 | 0.42 |
ENSMUST00000183358.1
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr13_-_64497792 | 0.42 |
ENSMUST00000180282.1
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene |
| chr14_+_66344369 | 0.41 |
ENSMUST00000118426.1
ENSMUST00000121955.1 ENSMUST00000120229.1 ENSMUST00000134440.1 |
Stmn4
|
stathmin-like 4 |
| chr2_+_48949495 | 0.41 |
ENSMUST00000112745.1
|
Mbd5
|
methyl-CpG binding domain protein 5 |
| chrX_+_56779437 | 0.40 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_+_66968559 | 0.39 |
ENSMUST00000127184.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr17_+_75435886 | 0.39 |
ENSMUST00000164192.1
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr1_+_75375271 | 0.39 |
ENSMUST00000087122.5
|
Speg
|
SPEG complex locus |
| chr2_+_27676440 | 0.38 |
ENSMUST00000129514.1
|
Rxra
|
retinoid X receptor alpha |
| chr15_-_89425856 | 0.38 |
ENSMUST00000109313.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr19_-_5802640 | 0.38 |
ENSMUST00000173523.1
ENSMUST00000173499.1 ENSMUST00000172812.2 |
Malat1
|
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
| chr14_-_54966925 | 0.37 |
ENSMUST00000111456.1
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr5_-_123141067 | 0.36 |
ENSMUST00000162697.1
ENSMUST00000160321.1 ENSMUST00000159637.1 |
AI480526
|
expressed sequence AI480526 |
| chr14_+_55575617 | 0.35 |
ENSMUST00000022826.5
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr14_-_57058030 | 0.35 |
ENSMUST00000061614.6
|
Gja3
|
gap junction protein, alpha 3 |
| chr3_-_142395661 | 0.34 |
ENSMUST00000029941.9
ENSMUST00000168967.2 ENSMUST00000090134.5 ENSMUST00000058626.8 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr1_+_63445842 | 0.33 |
ENSMUST00000087374.3
ENSMUST00000114107.1 ENSMUST00000182642.1 |
Adam23
|
a disintegrin and metallopeptidase domain 23 |
| chr8_-_41016749 | 0.33 |
ENSMUST00000117735.1
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr1_-_64122256 | 0.33 |
ENSMUST00000135075.1
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr19_-_24861828 | 0.32 |
ENSMUST00000047666.4
|
Pgm5
|
phosphoglucomutase 5 |
| chr9_+_74953053 | 0.32 |
ENSMUST00000170846.1
|
Fam214a
|
family with sequence similarity 214, member A |
| chr16_+_11313812 | 0.32 |
ENSMUST00000023140.5
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr13_-_59556845 | 0.31 |
ENSMUST00000170378.1
ENSMUST00000169434.1 |
Agtpbp1
|
ATP/GTP binding protein 1 |
| chr15_-_76232045 | 0.30 |
ENSMUST00000167754.1
|
Plec
|
plectin |
| chr15_-_89425795 | 0.30 |
ENSMUST00000168376.1
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr9_-_7873157 | 0.30 |
ENSMUST00000159323.1
ENSMUST00000115673.2 |
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr16_+_7069825 | 0.30 |
ENSMUST00000056416.7
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr18_-_38918642 | 0.30 |
ENSMUST00000040647.4
|
Fgf1
|
fibroblast growth factor 1 |
| chr14_-_50897456 | 0.30 |
ENSMUST00000170855.1
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr10_+_63457505 | 0.29 |
ENSMUST00000105440.1
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr4_-_55532453 | 0.29 |
ENSMUST00000132746.1
ENSMUST00000107619.2 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr4_-_83021102 | 0.28 |
ENSMUST00000071708.5
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr5_-_17888884 | 0.28 |
ENSMUST00000169095.1
|
Cd36
|
CD36 antigen |
| chr1_-_134234492 | 0.28 |
ENSMUST00000169927.1
|
Adora1
|
adenosine A1 receptor |
| chr12_+_69241832 | 0.27 |
ENSMUST00000063445.6
|
Klhdc1
|
kelch domain containing 1 |
| chr8_+_81342556 | 0.27 |
ENSMUST00000172167.1
ENSMUST00000169116.1 ENSMUST00000109852.2 ENSMUST00000172031.1 |
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chrX_+_6577259 | 0.27 |
ENSMUST00000089520.2
|
Shroom4
|
shroom family member 4 |
| chr8_+_40307458 | 0.27 |
ENSMUST00000068999.7
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr12_+_109747903 | 0.26 |
ENSMUST00000183084.1
ENSMUST00000182300.1 |
Mirg
|
miRNA containing gene |
| chr1_+_180109192 | 0.25 |
ENSMUST00000143176.1
ENSMUST00000135056.1 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr5_+_25759987 | 0.24 |
ENSMUST00000128727.1
ENSMUST00000088244.4 |
Actr3b
|
ARP3 actin-related protein 3B |
| chr6_+_108213086 | 0.24 |
ENSMUST00000032192.6
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr5_-_113081579 | 0.24 |
ENSMUST00000131708.1
ENSMUST00000117143.1 ENSMUST00000119627.1 |
Crybb3
|
crystallin, beta B3 |
| chr17_+_8283762 | 0.24 |
ENSMUST00000155364.1
ENSMUST00000046754.8 ENSMUST00000124023.1 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr3_-_149185495 | 0.24 |
ENSMUST00000066092.3
|
Gm9912
|
predicted gene 9912 |
| chr3_+_51693771 | 0.23 |
ENSMUST00000099104.2
|
Gm10729
|
predicted gene 10729 |
| chr5_-_24540976 | 0.23 |
ENSMUST00000117900.1
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr5_+_66968416 | 0.22 |
ENSMUST00000038188.7
|
Limch1
|
LIM and calponin homology domains 1 |
| chr14_-_93888732 | 0.22 |
ENSMUST00000068992.2
|
Pcdh9
|
protocadherin 9 |
| chr6_+_21986887 | 0.22 |
ENSMUST00000151315.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_-_71262232 | 0.22 |
ENSMUST00000129630.2
ENSMUST00000114186.2 ENSMUST00000074301.3 |
Smyd1
|
SET and MYND domain containing 1 |
| chr3_+_138143429 | 0.22 |
ENSMUST00000040321.6
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr13_-_85127514 | 0.21 |
ENSMUST00000179230.1
|
Gm4076
|
predicted gene 4076 |
| chr12_+_59130994 | 0.20 |
ENSMUST00000177460.1
|
Ctage5
|
CTAGE family, member 5 |
| chr8_-_61902669 | 0.20 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr17_+_44188564 | 0.19 |
ENSMUST00000024755.5
|
Clic5
|
chloride intracellular channel 5 |
| chr6_-_14755250 | 0.19 |
ENSMUST00000045096.4
|
Ppp1r3a
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A |
| chr13_+_102693522 | 0.19 |
ENSMUST00000022124.3
ENSMUST00000171267.1 ENSMUST00000167144.1 ENSMUST00000170878.1 |
Cd180
|
CD180 antigen |
| chr10_+_69785507 | 0.18 |
ENSMUST00000182993.1
|
Ank3
|
ankyrin 3, epithelial |
| chr1_-_88702121 | 0.18 |
ENSMUST00000159814.1
|
Arl4c
|
ADP-ribosylation factor-like 4C |
| chr3_-_56183678 | 0.18 |
ENSMUST00000029374.6
|
Nbea
|
neurobeachin |
| chr13_-_107890059 | 0.16 |
ENSMUST00000105097.2
|
Zswim6
|
zinc finger SWIM-type containing 6 |
| chrX_+_164373363 | 0.15 |
ENSMUST00000033751.7
|
Figf
|
c-fos induced growth factor |
| chr9_-_66514567 | 0.15 |
ENSMUST00000056890.8
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr6_+_42286709 | 0.14 |
ENSMUST00000163936.1
|
Clcn1
|
chloride channel 1 |
| chr18_+_45268876 | 0.14 |
ENSMUST00000183850.1
ENSMUST00000066890.7 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr7_-_67372846 | 0.14 |
ENSMUST00000156690.1
ENSMUST00000107476.1 ENSMUST00000076325.5 ENSMUST00000032776.8 ENSMUST00000133074.1 |
Mef2a
|
myocyte enhancer factor 2A |
| chr5_+_107497718 | 0.14 |
ENSMUST00000112671.2
|
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr9_+_55326913 | 0.14 |
ENSMUST00000085754.3
ENSMUST00000034862.4 |
AI118078
|
expressed sequence AI118078 |
| chr3_-_49757257 | 0.14 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr16_+_30065333 | 0.12 |
ENSMUST00000023171.7
|
Hes1
|
hairy and enhancer of split 1 (Drosophila) |
| chr5_+_65348386 | 0.12 |
ENSMUST00000031096.7
|
Klb
|
klotho beta |
| chr3_-_51340628 | 0.12 |
ENSMUST00000062009.7
|
Elf2
|
E74-like factor 2 |
| chr7_+_30121915 | 0.11 |
ENSMUST00000098596.3
ENSMUST00000153792.1 |
Zfp382
|
zinc finger protein 382 |
| chr11_+_3330401 | 0.11 |
ENSMUST00000045153.4
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_+_27165233 | 0.11 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr17_+_12378603 | 0.10 |
ENSMUST00000014578.5
|
Plg
|
plasminogen |
| chr16_+_45093611 | 0.10 |
ENSMUST00000099498.2
|
Ccdc80
|
coiled-coil domain containing 80 |
| chrX_-_47892396 | 0.10 |
ENSMUST00000153548.2
|
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr4_+_123201503 | 0.09 |
ENSMUST00000068262.5
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr4_+_133553370 | 0.08 |
ENSMUST00000042706.2
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr8_-_124569696 | 0.08 |
ENSMUST00000063278.6
|
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 1.2 | 3.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.1 | 7.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.9 | 4.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.8 | 2.5 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.8 | 3.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.7 | 7.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.5 | 2.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.5 | 4.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.5 | 5.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 2.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 | 3.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 19.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 0.9 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 0.9 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 0.8 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.3 | 0.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 1.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 6.8 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.2 | 14.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.2 | 0.8 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 0.6 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 1.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 4.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.2 | 0.6 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.4 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 | 0.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 0.3 | GO:0070256 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 3.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.4 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 1.2 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 4.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.9 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 4.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.5 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0045168 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) cell-cell signaling involved in cell fate commitment(GO:0045168) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.2 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.4 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.9 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.7 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.8 | GO:0003012 | muscle system process(GO:0003012) |
| 0.0 | 3.2 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.1 | 20.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.8 | 7.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 2.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.4 | 10.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 3.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 2.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 5.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 3.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 1.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 32.3 | GO:0031674 | I band(GO:0031674) |
| 0.2 | 0.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 6.0 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 7.1 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 0.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.0 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.3 | 7.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 1.1 | 10.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 15.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 3.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 3.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 3.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 2.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 11.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 3.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.7 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 2.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 2.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 4.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0071820 | N-box binding(GO:0071820) |
| 0.0 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 1.5 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 14.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 8.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.0 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.0 | 7.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 43.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 3.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |