Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
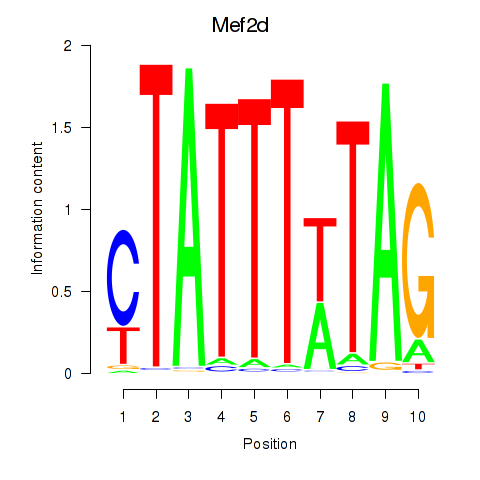
Results for Mef2d_Mef2a
Z-value: 1.99
Transcription factors associated with Mef2d_Mef2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2d
|
ENSMUSG00000001419.11 | myocyte enhancer factor 2D |
|
Mef2a
|
ENSMUSG00000030557.10 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2d | mm10_v2_chr3_+_88142328_88142483 | 0.21 | 2.1e-01 | Click! |
| Mef2a | mm10_v2_chr7_-_67372846_67372858 | 0.10 | 5.5e-01 | Click! |
Activity profile of Mef2d_Mef2a motif
Sorted Z-values of Mef2d_Mef2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_66935333 | 19.05 |
ENSMUST00000120415.1
ENSMUST00000119429.1 |
Myl1
|
myosin, light polypeptide 1 |
| chr11_+_98383811 | 16.14 |
ENSMUST00000008021.2
|
Tcap
|
titin-cap |
| chr5_+_122100951 | 12.56 |
ENSMUST00000014080.6
ENSMUST00000111750.1 ENSMUST00000139213.1 ENSMUST00000111751.1 ENSMUST00000155612.1 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr3_-_123034943 | 11.67 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr7_+_142441808 | 11.14 |
ENSMUST00000105971.1
ENSMUST00000145287.1 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chrX_+_157699113 | 10.86 |
ENSMUST00000112521.1
|
Smpx
|
small muscle protein, X-linked |
| chr8_+_15057646 | 10.39 |
ENSMUST00000033842.3
|
Myom2
|
myomesin 2 |
| chr1_-_66945019 | 10.13 |
ENSMUST00000027151.5
|
Myl1
|
myosin, light polypeptide 1 |
| chr1_+_40805578 | 9.56 |
ENSMUST00000114765.2
|
Tmem182
|
transmembrane protein 182 |
| chr14_-_34588654 | 9.29 |
ENSMUST00000022328.6
ENSMUST00000064098.6 ENSMUST00000022327.5 ENSMUST00000022330.7 |
Ldb3
|
LIM domain binding 3 |
| chr1_-_66945361 | 9.27 |
ENSMUST00000160100.1
|
Myl1
|
myosin, light polypeptide 1 |
| chr3_+_103074009 | 9.01 |
ENSMUST00000090715.6
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr11_+_67277124 | 9.01 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr11_-_94976327 | 8.20 |
ENSMUST00000103162.1
ENSMUST00000166320.1 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr11_+_67798269 | 7.96 |
ENSMUST00000168612.1
ENSMUST00000040574.4 |
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr7_-_126463100 | 7.95 |
ENSMUST00000032974.6
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr10_+_33083476 | 7.57 |
ENSMUST00000095762.4
|
Trdn
|
triadin |
| chr11_+_67200052 | 7.33 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr14_-_21714570 | 7.30 |
ENSMUST00000073870.5
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr3_+_103074035 | 6.82 |
ENSMUST00000176440.1
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr7_-_128206346 | 6.58 |
ENSMUST00000033049.7
|
Cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chrX_-_95478107 | 6.56 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr14_-_34588607 | 6.45 |
ENSMUST00000090040.4
|
Ldb3
|
LIM domain binding 3 |
| chr1_-_172219715 | 6.06 |
ENSMUST00000170700.1
ENSMUST00000003554.4 |
Casq1
|
calsequestrin 1 |
| chr14_-_20668269 | 5.65 |
ENSMUST00000057090.5
ENSMUST00000117386.1 |
Synpo2l
|
synaptopodin 2-like |
| chrX_+_157698910 | 5.61 |
ENSMUST00000136141.1
|
Smpx
|
small muscle protein, X-linked |
| chr15_-_77022632 | 5.50 |
ENSMUST00000019037.8
ENSMUST00000169226.1 |
Mb
|
myoglobin |
| chr7_+_142498832 | 5.38 |
ENSMUST00000078497.8
ENSMUST00000105953.3 ENSMUST00000179658.1 ENSMUST00000105954.3 ENSMUST00000105952.3 ENSMUST00000105955.1 ENSMUST00000074187.6 ENSMUST00000180152.1 ENSMUST00000105950.4 ENSMUST00000105957.3 ENSMUST00000169299.2 ENSMUST00000105958.3 ENSMUST00000105949.1 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr15_-_78206391 | 5.33 |
ENSMUST00000120592.1
|
Pvalb
|
parvalbumin |
| chr9_-_31913462 | 5.28 |
ENSMUST00000116615.3
|
Barx2
|
BarH-like homeobox 2 |
| chrX_+_101449078 | 5.13 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr3_-_123236134 | 5.09 |
ENSMUST00000106427.1
ENSMUST00000106426.1 ENSMUST00000051443.5 |
Synpo2
|
synaptopodin 2 |
| chr7_+_142442330 | 5.04 |
ENSMUST00000149529.1
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr13_-_113663670 | 4.98 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr7_+_102101736 | 4.97 |
ENSMUST00000033300.2
|
Art1
|
ADP-ribosyltransferase 1 |
| chr9_-_120023558 | 4.88 |
ENSMUST00000111635.2
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr4_-_73950834 | 4.58 |
ENSMUST00000095023.1
ENSMUST00000030101.3 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr10_-_88605017 | 4.49 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr3_+_102086471 | 4.44 |
ENSMUST00000165540.2
ENSMUST00000164123.1 |
Casq2
|
calsequestrin 2 |
| chr11_+_67238017 | 4.39 |
ENSMUST00000018632.4
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr3_+_102086415 | 4.38 |
ENSMUST00000029454.5
|
Casq2
|
calsequestrin 2 |
| chr9_+_121777607 | 4.21 |
ENSMUST00000098272.2
|
Klhl40
|
kelch-like 40 |
| chr5_-_112896350 | 4.20 |
ENSMUST00000086617.4
|
Myo18b
|
myosin XVIIIb |
| chr7_+_128003911 | 4.08 |
ENSMUST00000106248.1
|
Trim72
|
tripartite motif-containing 72 |
| chr14_-_20664562 | 3.87 |
ENSMUST00000119483.1
|
Synpo2l
|
synaptopodin 2-like |
| chr11_+_67200137 | 3.60 |
ENSMUST00000129018.1
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr14_-_54966570 | 3.44 |
ENSMUST00000124930.1
ENSMUST00000134256.1 ENSMUST00000081857.6 ENSMUST00000145322.1 |
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr19_-_56389877 | 3.35 |
ENSMUST00000166203.1
ENSMUST00000167239.1 ENSMUST00000040711.8 ENSMUST00000095947.4 ENSMUST00000073536.6 |
Nrap
|
nebulin-related anchoring protein |
| chr10_+_127501672 | 3.34 |
ENSMUST00000160019.1
ENSMUST00000160610.1 |
Stac3
|
SH3 and cysteine rich domain 3 |
| chr7_+_81057589 | 3.31 |
ENSMUST00000107348.1
|
Alpk3
|
alpha-kinase 3 |
| chr4_+_156215920 | 3.27 |
ENSMUST00000105572.1
|
2310042D19Rik
|
RIKEN cDNA 2310042D19 gene |
| chr16_-_36990449 | 3.25 |
ENSMUST00000075869.6
|
Fbxo40
|
F-box protein 40 |
| chrX_+_164438039 | 3.15 |
ENSMUST00000033755.5
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr6_+_42286676 | 3.10 |
ENSMUST00000031894.6
|
Clcn1
|
chloride channel 1 |
| chr9_+_110763646 | 3.04 |
ENSMUST00000079784.7
|
Myl3
|
myosin, light polypeptide 3 |
| chr11_+_70657196 | 2.99 |
ENSMUST00000157027.1
ENSMUST00000072841.5 ENSMUST00000108548.1 ENSMUST00000126241.1 |
Eno3
|
enolase 3, beta muscle |
| chr11_+_70657687 | 2.94 |
ENSMUST00000134087.1
ENSMUST00000170716.1 |
Eno3
|
enolase 3, beta muscle |
| chr1_+_135799402 | 2.85 |
ENSMUST00000152208.1
ENSMUST00000152075.1 ENSMUST00000154463.1 ENSMUST00000139986.1 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr17_-_48432723 | 2.81 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr17_+_71019548 | 2.73 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr14_-_32685246 | 2.73 |
ENSMUST00000096038.3
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene |
| chr10_+_127501707 | 2.71 |
ENSMUST00000035839.2
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr1_+_135799833 | 2.70 |
ENSMUST00000148201.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr2_+_32628390 | 2.70 |
ENSMUST00000156578.1
|
Ak1
|
adenylate kinase 1 |
| chr17_+_71019503 | 2.69 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chrX_+_133850980 | 2.60 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr2_-_76982455 | 2.58 |
ENSMUST00000011934.5
ENSMUST00000099981.2 ENSMUST00000099980.3 ENSMUST00000111882.2 ENSMUST00000140091.1 |
Ttn
|
titin |
| chr7_-_4514558 | 2.55 |
ENSMUST00000163538.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr2_+_32625431 | 2.49 |
ENSMUST00000113277.1
|
Ak1
|
adenylate kinase 1 |
| chr2_+_67445994 | 2.48 |
ENSMUST00000112347.1
ENSMUST00000028410.3 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr2_+_69670100 | 2.42 |
ENSMUST00000100050.3
|
Klhl41
|
kelch-like 41 |
| chr5_+_122101146 | 2.39 |
ENSMUST00000147178.1
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr13_-_93144557 | 2.38 |
ENSMUST00000062122.3
|
Cmya5
|
cardiomyopathy associated 5 |
| chr11_-_120648104 | 2.35 |
ENSMUST00000026134.2
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr11_+_77462325 | 2.20 |
ENSMUST00000102493.1
|
Coro6
|
coronin 6 |
| chr6_-_14755250 | 2.17 |
ENSMUST00000045096.4
|
Ppp1r3a
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A |
| chr7_-_4514609 | 2.14 |
ENSMUST00000166959.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr14_-_57058030 | 2.13 |
ENSMUST00000061614.6
|
Gja3
|
gap junction protein, alpha 3 |
| chr10_-_107494719 | 2.11 |
ENSMUST00000044210.3
|
Myf6
|
myogenic factor 6 |
| chr5_+_135887905 | 2.10 |
ENSMUST00000005077.6
|
Hspb1
|
heat shock protein 1 |
| chr5_+_135887988 | 2.09 |
ENSMUST00000111155.1
|
Hspb1
|
heat shock protein 1 |
| chr2_+_91118136 | 1.99 |
ENSMUST00000169776.1
ENSMUST00000111430.3 ENSMUST00000137942.1 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr9_+_77636494 | 1.96 |
ENSMUST00000057781.7
|
Klhl31
|
kelch-like 31 |
| chr3_-_94473591 | 1.89 |
ENSMUST00000029785.3
|
Riiad1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
| chr9_-_77347787 | 1.82 |
ENSMUST00000184848.1
ENSMUST00000184415.1 |
Mlip
|
muscular LMNA-interacting protein |
| chr16_-_4523056 | 1.67 |
ENSMUST00000090500.3
ENSMUST00000023161.7 |
Srl
|
sarcalumenin |
| chr3_+_32817520 | 1.67 |
ENSMUST00000072312.5
ENSMUST00000108228.1 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr1_+_134193432 | 1.63 |
ENSMUST00000038445.6
|
Mybph
|
myosin binding protein H |
| chr6_+_42286709 | 1.62 |
ENSMUST00000163936.1
|
Clcn1
|
chloride channel 1 |
| chr7_+_45405960 | 1.60 |
ENSMUST00000107774.1
|
Kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr19_+_6384395 | 1.60 |
ENSMUST00000035269.8
ENSMUST00000113483.1 |
Pygm
|
muscle glycogen phosphorylase |
| chr9_-_49798905 | 1.59 |
ENSMUST00000114476.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr4_+_134315112 | 1.58 |
ENSMUST00000105875.1
ENSMUST00000030638.6 |
Trim63
|
tripartite motif-containing 63 |
| chr9_-_77347816 | 1.56 |
ENSMUST00000184138.1
ENSMUST00000184006.1 ENSMUST00000185144.1 ENSMUST00000034910.9 |
Mlip
|
muscular LMNA-interacting protein |
| chr2_+_27165233 | 1.54 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr5_-_24540439 | 1.48 |
ENSMUST00000048302.6
ENSMUST00000119657.1 |
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr15_-_89425795 | 1.45 |
ENSMUST00000168376.1
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr1_-_36535719 | 1.45 |
ENSMUST00000054665.4
|
Ankrd23
|
ankyrin repeat domain 23 |
| chr1_+_75375271 | 1.44 |
ENSMUST00000087122.5
|
Speg
|
SPEG complex locus |
| chr7_-_4514368 | 1.42 |
ENSMUST00000166161.1
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr3_+_138065052 | 1.37 |
ENSMUST00000163080.2
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr10_+_33863935 | 1.36 |
ENSMUST00000092597.3
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr6_+_127015542 | 1.36 |
ENSMUST00000000187.5
|
Fgf6
|
fibroblast growth factor 6 |
| chr1_-_172297989 | 1.33 |
ENSMUST00000085913.4
ENSMUST00000097464.2 ENSMUST00000137679.1 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr7_-_48848023 | 1.24 |
ENSMUST00000032658.6
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr15_+_25940846 | 1.24 |
ENSMUST00000110438.1
|
Fam134b
|
family with sequence similarity 134, member B |
| chr19_-_45235811 | 1.22 |
ENSMUST00000099401.4
|
Lbx1
|
ladybird homeobox homolog 1 (Drosophila) |
| chr4_-_99654983 | 1.17 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr6_-_71262232 | 1.16 |
ENSMUST00000129630.2
ENSMUST00000114186.2 ENSMUST00000074301.3 |
Smyd1
|
SET and MYND domain containing 1 |
| chr9_-_77347889 | 1.15 |
ENSMUST00000185039.1
|
Mlip
|
muscular LMNA-interacting protein |
| chr7_-_102100227 | 1.13 |
ENSMUST00000106937.1
|
Art5
|
ADP-ribosyltransferase 5 |
| chr7_-_97417730 | 1.09 |
ENSMUST00000043077.7
|
Thrsp
|
thyroid hormone responsive |
| chr14_+_26894557 | 1.07 |
ENSMUST00000090337.4
ENSMUST00000165929.2 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr19_-_53464721 | 1.01 |
ENSMUST00000180489.1
|
5830416P10Rik
|
RIKEN cDNA 5830416P10 gene |
| chr1_-_74748577 | 0.98 |
ENSMUST00000113672.2
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catatlytic subunit |
| chr12_-_109600328 | 0.97 |
ENSMUST00000149046.2
|
Rtl1
|
retrotransposon-like 1 |
| chr15_-_89425856 | 0.96 |
ENSMUST00000109313.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr9_-_49798729 | 0.92 |
ENSMUST00000166811.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr1_+_75360321 | 0.92 |
ENSMUST00000027409.9
|
Des
|
desmin |
| chr13_-_64497792 | 0.91 |
ENSMUST00000180282.1
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene |
| chr15_+_94629148 | 0.91 |
ENSMUST00000080141.4
|
Tmem117
|
transmembrane protein 117 |
| chr16_+_78930940 | 0.89 |
ENSMUST00000114216.1
ENSMUST00000069148.6 ENSMUST00000023568.7 |
Chodl
|
chondrolectin |
| chrX_+_73774397 | 0.88 |
ENSMUST00000002081.5
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr12_+_52699297 | 0.88 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_187608755 | 0.88 |
ENSMUST00000127489.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr4_-_4138432 | 0.87 |
ENSMUST00000070375.7
|
Penk
|
preproenkephalin |
| chrX_-_47892432 | 0.86 |
ENSMUST00000141084.2
|
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr11_-_53618659 | 0.86 |
ENSMUST00000000889.6
|
Il4
|
interleukin 4 |
| chr3_-_88384433 | 0.82 |
ENSMUST00000076048.4
|
Bglap
|
bone gamma carboxyglutamate protein |
| chr14_-_101729690 | 0.81 |
ENSMUST00000066461.3
|
Gm9922
|
predicted gene 9922 |
| chr1_+_70725902 | 0.80 |
ENSMUST00000161937.1
ENSMUST00000162182.1 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr7_-_81566939 | 0.79 |
ENSMUST00000042318.5
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr17_+_85028347 | 0.78 |
ENSMUST00000024944.7
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr3_+_19644452 | 0.77 |
ENSMUST00000029139.7
|
Trim55
|
tripartite motif-containing 55 |
| chr18_+_60963517 | 0.76 |
ENSMUST00000115295.2
ENSMUST00000039904.6 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_+_102268732 | 0.74 |
ENSMUST00000036467.4
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr1_-_74749221 | 0.73 |
ENSMUST00000081636.6
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catatlytic subunit |
| chr1_-_160153571 | 0.73 |
ENSMUST00000039178.5
|
Tnn
|
tenascin N |
| chrX_-_47892396 | 0.73 |
ENSMUST00000153548.2
|
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr1_+_70725715 | 0.72 |
ENSMUST00000053922.5
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr5_+_92386879 | 0.71 |
ENSMUST00000128246.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr1_-_163725123 | 0.71 |
ENSMUST00000159679.1
|
Mettl11b
|
methyltransferase like 11B |
| chr2_+_36216749 | 0.70 |
ENSMUST00000147012.1
ENSMUST00000122948.1 |
Gm13431
|
predicted gene 13431 |
| chr16_+_7069825 | 0.70 |
ENSMUST00000056416.7
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr18_+_67464849 | 0.68 |
ENSMUST00000025411.7
|
Slmo1
|
slowmo homolog 1 (Drosophila) |
| chr12_-_112929415 | 0.66 |
ENSMUST00000075827.3
|
Jag2
|
jagged 2 |
| chr2_+_155611175 | 0.66 |
ENSMUST00000092995.5
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr7_-_126800036 | 0.66 |
ENSMUST00000133514.1
ENSMUST00000151137.1 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr11_-_69948145 | 0.65 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr13_+_83573577 | 0.65 |
ENSMUST00000185052.1
|
Mef2c
|
myocyte enhancer factor 2C |
| chrX_-_165004829 | 0.63 |
ENSMUST00000114890.2
|
Gm17604
|
predicted gene, 17604 |
| chr6_-_55133014 | 0.62 |
ENSMUST00000003568.8
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr6_-_55132841 | 0.61 |
ENSMUST00000164012.1
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr15_+_103013815 | 0.61 |
ENSMUST00000001709.2
|
Hoxc5
|
homeobox C5 |
| chr11_+_3330401 | 0.61 |
ENSMUST00000045153.4
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr15_+_62178175 | 0.60 |
ENSMUST00000182476.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr13_-_23683941 | 0.59 |
ENSMUST00000171127.1
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr12_+_108334341 | 0.57 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr2_-_120289063 | 0.57 |
ENSMUST00000094665.4
|
Pla2g4d
|
phospholipase A2, group IVD |
| chr11_-_107716517 | 0.56 |
ENSMUST00000021065.5
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr4_-_134372529 | 0.55 |
ENSMUST00000030643.2
|
Extl1
|
exostoses (multiple)-like 1 |
| chr7_-_126800354 | 0.54 |
ENSMUST00000106348.1
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr6_-_88627422 | 0.53 |
ENSMUST00000120933.2
ENSMUST00000169512.1 |
Kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chrX_-_106840572 | 0.52 |
ENSMUST00000062010.9
|
Zcchc5
|
zinc finger, CCHC domain containing 5 |
| chr13_-_21753851 | 0.51 |
ENSMUST00000074752.2
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr2_-_25470031 | 0.51 |
ENSMUST00000114251.1
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr5_+_66745835 | 0.50 |
ENSMUST00000101164.4
ENSMUST00000118242.1 ENSMUST00000119854.1 ENSMUST00000117601.1 |
Limch1
|
LIM and calponin homology domains 1 |
| chr7_+_99735145 | 0.50 |
ENSMUST00000098264.1
|
Olfr520
|
olfactory receptor 520 |
| chr15_-_95528228 | 0.50 |
ENSMUST00000075275.2
|
Nell2
|
NEL-like 2 |
| chr14_-_54966925 | 0.49 |
ENSMUST00000111456.1
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr4_+_46039202 | 0.48 |
ENSMUST00000156200.1
|
Tmod1
|
tropomodulin 1 |
| chr19_-_60790692 | 0.46 |
ENSMUST00000025955.6
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr2_-_25469742 | 0.45 |
ENSMUST00000114259.2
ENSMUST00000015234.6 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr9_-_112187898 | 0.45 |
ENSMUST00000178410.1
ENSMUST00000172380.3 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr3_-_95882232 | 0.44 |
ENSMUST00000161866.1
|
Gm129
|
predicted gene 129 |
| chr1_-_86111970 | 0.43 |
ENSMUST00000027431.6
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr3_-_95882193 | 0.43 |
ENSMUST00000159863.1
ENSMUST00000159739.1 ENSMUST00000036418.3 |
Gm129
|
predicted gene 129 |
| chr1_-_172329261 | 0.43 |
ENSMUST00000062387.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr19_+_38264761 | 0.42 |
ENSMUST00000087252.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr8_+_31089471 | 0.41 |
ENSMUST00000036631.7
ENSMUST00000170204.1 |
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr2_+_105682463 | 0.40 |
ENSMUST00000140173.1
ENSMUST00000135412.1 ENSMUST00000138365.1 ENSMUST00000145744.1 |
Pax6
|
paired box gene 6 |
| chr12_-_111980751 | 0.39 |
ENSMUST00000170525.1
|
BC048943
|
cDNA sequence BC048943 |
| chr11_+_95712673 | 0.39 |
ENSMUST00000107717.1
|
Zfp652
|
zinc finger protein 652 |
| chr6_+_5390387 | 0.39 |
ENSMUST00000183358.1
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr2_+_135659625 | 0.38 |
ENSMUST00000134310.1
|
Plcb4
|
phospholipase C, beta 4 |
| chr19_+_47228804 | 0.38 |
ENSMUST00000111807.3
|
Neurl1a
|
neuralized homolog 1A (Drosophila) |
| chr2_+_129198757 | 0.37 |
ENSMUST00000028880.3
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr2_+_61578549 | 0.35 |
ENSMUST00000112502.1
ENSMUST00000078074.2 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr5_-_123140135 | 0.33 |
ENSMUST00000160099.1
|
AI480526
|
expressed sequence AI480526 |
| chr11_+_111066154 | 0.32 |
ENSMUST00000042970.2
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr18_+_76059458 | 0.32 |
ENSMUST00000167921.1
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr15_-_36879816 | 0.32 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr11_+_120949053 | 0.30 |
ENSMUST00000154187.1
ENSMUST00000100130.3 ENSMUST00000129473.1 ENSMUST00000168579.1 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr16_+_32271468 | 0.29 |
ENSMUST00000093183.3
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr19_-_31765027 | 0.29 |
ENSMUST00000065067.6
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr9_-_105521147 | 0.29 |
ENSMUST00000176770.1
ENSMUST00000085133.6 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr17_+_34039437 | 0.29 |
ENSMUST00000131134.1
ENSMUST00000087497.4 ENSMUST00000114255.1 ENSMUST00000114252.1 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr2_-_104028287 | 0.28 |
ENSMUST00000056170.3
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr10_-_81202037 | 0.28 |
ENSMUST00000005069.6
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr3_+_30746325 | 0.28 |
ENSMUST00000108262.3
|
Samd7
|
sterile alpha motif domain containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2d_Mef2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 2.8 | 8.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 2.5 | 7.6 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 2.4 | 23.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 2.1 | 14.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 2.0 | 6.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.8 | 8.8 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 1.8 | 15.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.3 | 8.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 1.2 | 6.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.9 | 41.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.9 | 5.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.8 | 4.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.8 | 2.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.8 | 1.6 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 5.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.7 | 8.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.7 | 1.3 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.6 | 5.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.6 | 1.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.6 | 4.2 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.6 | 1.7 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.6 | 1.7 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.5 | 5.4 | GO:0042637 | catagen(GO:0042637) |
| 0.5 | 1.5 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.5 | 2.4 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.5 | 4.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 6.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 34.0 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.4 | 0.9 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.4 | 2.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 3.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 56.9 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.3 | 1.2 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.3 | 0.9 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.3 | 2.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 4.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 0.6 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.2 | 0.7 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 4.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.2 | 6.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 4.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 0.4 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 2.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 9.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.7 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 10.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.4 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 1.6 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.3 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.8 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 1.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.3 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 2.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 4.7 | GO:0003012 | muscle system process(GO:0003012) |
| 0.0 | 2.1 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 1.2 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.9 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 1.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 2.5 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.7 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 1.2 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 1.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.3 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 3.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 14.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.8 | 8.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 2.8 | 19.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 2.5 | 22.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.9 | 33.3 | GO:0005861 | troponin complex(GO:0005861) |
| 1.5 | 39.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.3 | 13.4 | GO:0032982 | myosin filament(GO:0032982) |
| 1.2 | 7.3 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 8.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.7 | 5.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 5.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 1.5 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.5 | 10.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.5 | 33.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.5 | 1.8 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 65.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.4 | 5.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 5.7 | GO:0031674 | I band(GO:0031674) |
| 0.3 | 5.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 0.8 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 6.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0000802 | transverse filament(GO:0000802) |
| 0.1 | 1.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 4.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0036379 | myofilament(GO:0036379) |
| 0.0 | 3.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 3.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 27.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 2.2 | 22.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.9 | 50.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.8 | 15.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.6 | 9.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.7 | 5.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.7 | 8.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.7 | 5.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 1.8 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.5 | 1.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.5 | 6.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.5 | 2.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 2.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 5.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 1.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.4 | 18.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 1.6 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 0.9 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.3 | 4.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 3.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 4.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 5.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.8 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 5.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 4.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 7.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 2.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.3 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 4.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 8.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 4.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 56.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.3 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 15.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 3.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 5.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 12.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.7 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 2.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 5.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 14.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 2.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 119.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.8 | 15.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.5 | 5.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.4 | 8.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 5.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 7.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 12.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |