Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
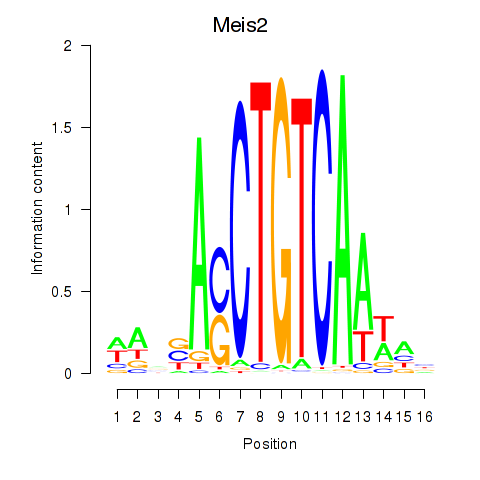
Results for Meis2
Z-value: 1.01
Transcription factors associated with Meis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meis2
|
ENSMUSG00000027210.14 | Meis homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis2 | mm10_v2_chr2_-_116064512_116064598 | 0.30 | 8.0e-02 | Click! |
Activity profile of Meis2 motif
Sorted Z-values of Meis2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_81975742 | 8.19 |
ENSMUST00000029645.8
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr19_-_8131982 | 5.73 |
ENSMUST00000065651.4
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr14_-_34355383 | 5.23 |
ENSMUST00000052126.5
|
Fam25c
|
family with sequence similarity 25, member C |
| chr1_+_88211956 | 4.99 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr1_-_121327672 | 4.30 |
ENSMUST00000159085.1
ENSMUST00000159125.1 ENSMUST00000161818.1 |
Insig2
|
insulin induced gene 2 |
| chr19_-_8405060 | 4.28 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr15_-_60921270 | 4.21 |
ENSMUST00000096418.3
|
A1bg
|
alpha-1-B glycoprotein |
| chr7_+_25897620 | 4.03 |
ENSMUST00000072438.6
ENSMUST00000005477.5 |
Cyp2b10
|
cytochrome P450, family 2, subfamily b, polypeptide 10 |
| chr2_+_177508570 | 4.02 |
ENSMUST00000108940.2
|
Gm14403
|
predicted gene 14403 |
| chr8_+_104733997 | 4.01 |
ENSMUST00000034346.8
ENSMUST00000164182.2 |
Ces2a
|
carboxylesterase 2A |
| chr3_+_107230608 | 3.85 |
ENSMUST00000179399.1
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr5_-_87337165 | 3.78 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr5_-_87254804 | 3.60 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr5_+_114175889 | 3.57 |
ENSMUST00000146841.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr1_-_121327734 | 3.52 |
ENSMUST00000160968.1
ENSMUST00000162582.1 |
Insig2
|
insulin induced gene 2 |
| chr15_+_4727265 | 3.44 |
ENSMUST00000162350.1
|
C6
|
complement component 6 |
| chr7_+_119617781 | 3.34 |
ENSMUST00000047929.6
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr6_-_85915604 | 3.25 |
ENSMUST00000174369.1
|
Cml1
|
camello-like 1 |
| chr7_+_119617804 | 3.24 |
ENSMUST00000135683.1
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr15_+_82555087 | 3.21 |
ENSMUST00000068861.6
|
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr15_+_4727175 | 3.18 |
ENSMUST00000162585.1
|
C6
|
complement component 6 |
| chr8_-_117673682 | 2.95 |
ENSMUST00000173522.1
ENSMUST00000174450.1 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr9_+_106448629 | 2.92 |
ENSMUST00000048527.7
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr10_+_107271827 | 2.91 |
ENSMUST00000020057.8
ENSMUST00000105280.3 |
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr3_+_19985612 | 2.78 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr8_-_117671526 | 2.77 |
ENSMUST00000037955.7
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr11_+_118443471 | 2.73 |
ENSMUST00000133558.1
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_-_27246814 | 2.72 |
ENSMUST00000149733.1
|
Sardh
|
sarcosine dehydrogenase |
| chr13_+_24845122 | 2.71 |
ENSMUST00000006893.8
|
D130043K22Rik
|
RIKEN cDNA D130043K22 gene |
| chr8_+_104847061 | 2.71 |
ENSMUST00000055052.5
|
Ces2c
|
carboxylesterase 2C |
| chr3_-_107969162 | 2.69 |
ENSMUST00000004136.8
ENSMUST00000106678.1 |
Gstm3
|
glutathione S-transferase, mu 3 |
| chr15_-_100599864 | 2.57 |
ENSMUST00000177247.2
ENSMUST00000177505.2 |
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr11_+_77518566 | 2.57 |
ENSMUST00000147386.1
|
Abhd15
|
abhydrolase domain containing 15 |
| chr10_+_40629987 | 2.55 |
ENSMUST00000019977.7
|
Ddo
|
D-aspartate oxidase |
| chr19_-_7966000 | 2.55 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr11_+_83746940 | 2.53 |
ENSMUST00000070832.2
|
1100001G20Rik
|
RIKEN cDNA 1100001G20 gene |
| chr4_+_141239499 | 2.53 |
ENSMUST00000141834.2
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr3_+_81999461 | 2.52 |
ENSMUST00000107736.1
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr14_-_66009204 | 2.52 |
ENSMUST00000059970.7
|
Gulo
|
gulonolactone (L-) oxidase |
| chr7_-_132576372 | 2.51 |
ENSMUST00000084500.6
|
Oat
|
ornithine aminotransferase |
| chr7_-_30944017 | 2.51 |
ENSMUST00000062620.7
|
Hamp
|
hepcidin antimicrobial peptide |
| chr6_-_71262232 | 2.50 |
ENSMUST00000129630.2
ENSMUST00000114186.2 ENSMUST00000074301.3 |
Smyd1
|
SET and MYND domain containing 1 |
| chr11_-_4118778 | 2.47 |
ENSMUST00000003681.7
|
Sec14l2
|
SEC14-like 2 (S. cerevisiae) |
| chr1_-_121327776 | 2.45 |
ENSMUST00000160688.1
|
Insig2
|
insulin induced gene 2 |
| chr6_-_106800051 | 2.45 |
ENSMUST00000013882.7
ENSMUST00000049675.4 ENSMUST00000113239.3 |
Crbn
|
cereblon |
| chr18_+_56432116 | 2.36 |
ENSMUST00000070166.5
|
Gramd3
|
GRAM domain containing 3 |
| chrX_+_59999436 | 2.35 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr16_+_22951072 | 2.34 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr1_+_167618246 | 2.33 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr15_+_4727202 | 2.32 |
ENSMUST00000161997.1
ENSMUST00000022788.8 |
C6
|
complement component 6 |
| chr11_-_50931612 | 2.31 |
ENSMUST00000109124.3
|
Zfp354b
|
zinc finger protein 354B |
| chr7_-_127273919 | 2.27 |
ENSMUST00000082428.3
|
Sephs2
|
selenophosphate synthetase 2 |
| chr18_+_31931470 | 2.27 |
ENSMUST00000025254.7
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr2_+_71981184 | 2.26 |
ENSMUST00000090826.5
ENSMUST00000102698.3 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_9547948 | 2.24 |
ENSMUST00000144177.1
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr7_+_44590886 | 2.11 |
ENSMUST00000107906.3
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr13_-_71963713 | 2.09 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr8_-_109579056 | 2.09 |
ENSMUST00000074898.6
|
Hp
|
haptoglobin |
| chr9_-_71168657 | 2.07 |
ENSMUST00000113570.1
|
Aqp9
|
aquaporin 9 |
| chr16_+_13903152 | 2.07 |
ENSMUST00000128757.1
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr1_+_88055377 | 2.06 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr5_-_34187670 | 2.05 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr5_+_105732063 | 2.02 |
ENSMUST00000154807.1
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr6_-_124636085 | 1.98 |
ENSMUST00000068797.2
|
Gm5077
|
predicted gene 5077 |
| chr1_-_52727457 | 1.98 |
ENSMUST00000156876.1
ENSMUST00000087701.3 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr7_-_126584578 | 1.95 |
ENSMUST00000150311.1
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr15_-_76232045 | 1.92 |
ENSMUST00000167754.1
|
Plec
|
plectin |
| chr2_+_27709247 | 1.91 |
ENSMUST00000100251.2
|
Rxra
|
retinoid X receptor alpha |
| chr1_+_87574016 | 1.86 |
ENSMUST00000166259.1
ENSMUST00000172222.1 ENSMUST00000163606.1 |
Neu2
|
neuraminidase 2 |
| chr1_-_180195902 | 1.85 |
ENSMUST00000161746.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr1_+_153899937 | 1.83 |
ENSMUST00000086199.5
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr1_-_153900198 | 1.81 |
ENSMUST00000123490.1
|
5830403L16Rik
|
RIKEN cDNA 5830403L16 gene |
| chr8_+_47824459 | 1.81 |
ENSMUST00000038693.6
|
Cldn22
|
claudin 22 |
| chr2_+_163122605 | 1.80 |
ENSMUST00000144092.1
|
Gm11454
|
predicted gene 11454 |
| chr11_+_77515104 | 1.79 |
ENSMUST00000094004.4
|
Abhd15
|
abhydrolase domain containing 15 |
| chr5_-_87424201 | 1.77 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr2_-_156004147 | 1.76 |
ENSMUST00000156993.1
ENSMUST00000141437.1 |
6430550D23Rik
|
RIKEN cDNA 6430550D23 gene |
| chr7_+_26173411 | 1.76 |
ENSMUST00000082214.4
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr11_-_94392917 | 1.75 |
ENSMUST00000178136.1
ENSMUST00000021231.7 |
Abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr15_-_77533312 | 1.75 |
ENSMUST00000062562.5
|
Apol7c
|
apolipoprotein L 7c |
| chr15_+_7129557 | 1.70 |
ENSMUST00000067190.5
ENSMUST00000164529.1 |
Lifr
|
leukemia inhibitory factor receptor |
| chr6_-_41035501 | 1.70 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr5_-_87569023 | 1.69 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr10_+_23894688 | 1.66 |
ENSMUST00000041416.7
|
Vnn1
|
vanin 1 |
| chr9_+_107542209 | 1.65 |
ENSMUST00000010201.3
|
Nprl2
|
nitrogen permease regulator-like 2 |
| chr16_-_23029012 | 1.64 |
ENSMUST00000039338.6
|
Kng2
|
kininogen 2 |
| chr11_-_51688614 | 1.62 |
ENSMUST00000007921.2
|
0610009B22Rik
|
RIKEN cDNA 0610009B22 gene |
| chr16_-_23029080 | 1.61 |
ENSMUST00000100046.2
|
Kng2
|
kininogen 2 |
| chr16_-_23029062 | 1.58 |
ENSMUST00000115349.2
|
Kng2
|
kininogen 2 |
| chr9_-_71771535 | 1.56 |
ENSMUST00000122065.1
ENSMUST00000121322.1 ENSMUST00000072899.2 |
Cgnl1
|
cingulin-like 1 |
| chr5_-_87140318 | 1.52 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr6_-_113435748 | 1.52 |
ENSMUST00000032416.4
ENSMUST00000113089.1 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr5_+_105731755 | 1.49 |
ENSMUST00000127686.1
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr5_+_92387673 | 1.48 |
ENSMUST00000145072.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr6_-_48445678 | 1.47 |
ENSMUST00000114556.1
|
Zfp467
|
zinc finger protein 467 |
| chrX_+_142228699 | 1.44 |
ENSMUST00000112913.1
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr15_+_10981747 | 1.43 |
ENSMUST00000070877.5
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr7_-_90475971 | 1.39 |
ENSMUST00000032843.7
|
Tmem126b
|
transmembrane protein 126B |
| chr9_-_106247730 | 1.39 |
ENSMUST00000112524.2
ENSMUST00000074082.6 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr7_-_126584220 | 1.38 |
ENSMUST00000128970.1
ENSMUST00000116269.2 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr1_+_194619815 | 1.37 |
ENSMUST00000027952.5
|
Plxna2
|
plexin A2 |
| chr13_+_114818232 | 1.35 |
ENSMUST00000166104.2
ENSMUST00000166176.2 ENSMUST00000184335.1 ENSMUST00000184245.1 ENSMUST00000015680.4 ENSMUST00000184214.1 ENSMUST00000165022.2 ENSMUST00000164737.1 ENSMUST00000184781.1 ENSMUST00000183407.1 ENSMUST00000184672.1 |
Mocs2
|
molybdenum cofactor synthesis 2 |
| chr8_-_41041828 | 1.33 |
ENSMUST00000051379.7
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chrX_+_36112110 | 1.32 |
ENSMUST00000033418.7
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr7_+_13398115 | 1.30 |
ENSMUST00000005791.7
|
Cabp5
|
calcium binding protein 5 |
| chr5_+_35814424 | 1.30 |
ENSMUST00000114203.1
|
Ablim2
|
actin-binding LIM protein 2 |
| chr4_-_118543210 | 1.29 |
ENSMUST00000156191.1
|
Tmem125
|
transmembrane protein 125 |
| chr9_+_108296853 | 1.28 |
ENSMUST00000035230.5
|
Amt
|
aminomethyltransferase |
| chr7_-_101581161 | 1.27 |
ENSMUST00000063920.2
|
Art2b
|
ADP-ribosyltransferase 2b |
| chr6_-_48445825 | 1.27 |
ENSMUST00000114561.2
|
Zfp467
|
zinc finger protein 467 |
| chr1_+_88055467 | 1.26 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr14_-_63987774 | 1.23 |
ENSMUST00000022532.5
|
4930578I06Rik
|
RIKEN cDNA 4930578I06 gene |
| chr12_-_103989950 | 1.23 |
ENSMUST00000120251.2
|
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr16_+_21891969 | 1.22 |
ENSMUST00000042065.6
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr5_+_138820080 | 1.22 |
ENSMUST00000179205.1
|
Gm5294
|
predicted gene 5294 |
| chr7_-_48456331 | 1.21 |
ENSMUST00000094384.3
|
Mrgprb1
|
MAS-related GPR, member B1 |
| chr2_-_30205794 | 1.21 |
ENSMUST00000113663.2
ENSMUST00000044038.3 |
Ccbl1
|
cysteine conjugate-beta lyase 1 |
| chr3_-_89387132 | 1.21 |
ENSMUST00000107433.1
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr6_-_48445373 | 1.21 |
ENSMUST00000114563.1
ENSMUST00000114558.1 ENSMUST00000101443.3 ENSMUST00000114564.1 |
Zfp467
|
zinc finger protein 467 |
| chr4_-_134372529 | 1.20 |
ENSMUST00000030643.2
|
Extl1
|
exostoses (multiple)-like 1 |
| chr1_-_184883218 | 1.20 |
ENSMUST00000048308.5
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr16_-_11909398 | 1.19 |
ENSMUST00000127972.1
ENSMUST00000121750.1 ENSMUST00000096272.4 ENSMUST00000073371.6 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr16_+_44173271 | 1.18 |
ENSMUST00000088356.4
ENSMUST00000169582.1 |
Gm608
|
predicted gene 608 |
| chr17_+_21423227 | 1.18 |
ENSMUST00000165230.1
ENSMUST00000007884.8 ENSMUST00000167749.1 |
Zfp54
|
zinc finger protein 54 |
| chr2_+_91710852 | 1.18 |
ENSMUST00000128140.1
ENSMUST00000140183.1 |
Harbi1
|
harbinger transposase derived 1 |
| chr2_-_30205772 | 1.17 |
ENSMUST00000113662.1
|
Ccbl1
|
cysteine conjugate-beta lyase 1 |
| chr16_+_23058250 | 1.17 |
ENSMUST00000039492.6
ENSMUST00000023589.8 ENSMUST00000089902.6 |
Kng1
|
kininogen 1 |
| chr8_-_3717547 | 1.17 |
ENSMUST00000058040.6
|
Gm9814
|
predicted gene 9814 |
| chr4_+_155562348 | 1.17 |
ENSMUST00000030939.7
|
Nadk
|
NAD kinase |
| chr6_-_33060172 | 1.17 |
ENSMUST00000115091.1
ENSMUST00000127666.1 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr9_+_123150941 | 1.16 |
ENSMUST00000026890.4
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr5_+_121660869 | 1.13 |
ENSMUST00000111765.1
|
Brap
|
BRCA1 associated protein |
| chr8_+_56294552 | 1.13 |
ENSMUST00000034026.8
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr4_-_32602760 | 1.13 |
ENSMUST00000056517.2
|
Gja10
|
gap junction protein, alpha 10 |
| chr7_-_126594941 | 1.12 |
ENSMUST00000058429.5
|
Il27
|
interleukin 27 |
| chr7_+_144175513 | 1.11 |
ENSMUST00000105900.1
|
Shank2
|
SH3/ankyrin domain gene 2 |
| chr3_+_107631322 | 1.11 |
ENSMUST00000106703.1
|
Gm10961
|
predicted gene 10961 |
| chr12_-_103989917 | 1.10 |
ENSMUST00000151709.2
ENSMUST00000176246.1 ENSMUST00000074693.7 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr5_-_72752763 | 1.09 |
ENSMUST00000113604.3
|
Txk
|
TXK tyrosine kinase |
| chr11_-_120617887 | 1.09 |
ENSMUST00000106188.3
ENSMUST00000026129.9 |
Pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr17_-_56717681 | 1.07 |
ENSMUST00000164907.1
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr1_-_52817503 | 1.07 |
ENSMUST00000162576.1
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr2_+_162987502 | 1.06 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr7_-_141010759 | 1.06 |
ENSMUST00000026565.6
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr5_-_138619653 | 1.06 |
ENSMUST00000129832.1
|
Zfp68
|
zinc finger protein 68 |
| chr6_-_142278836 | 1.04 |
ENSMUST00000111825.3
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr14_-_16249675 | 1.04 |
ENSMUST00000022311.4
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr13_+_119836006 | 1.04 |
ENSMUST00000179131.1
|
D13Ertd608e
|
DNA segment, Chr 13, ERATO Doi 608, expressed |
| chr5_-_107869153 | 1.04 |
ENSMUST00000128723.1
ENSMUST00000124034.1 |
Evi5
|
ecotropic viral integration site 5 |
| chr7_+_48959089 | 1.04 |
ENSMUST00000183659.1
|
Nav2
|
neuron navigator 2 |
| chr16_-_13903051 | 1.03 |
ENSMUST00000115803.1
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr16_+_44173239 | 1.02 |
ENSMUST00000119746.1
|
Gm608
|
predicted gene 608 |
| chr6_-_33060256 | 1.02 |
ENSMUST00000066379.4
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chrX_+_71364901 | 1.01 |
ENSMUST00000132837.1
|
Mtmr1
|
myotubularin related protein 1 |
| chr2_+_120977017 | 1.01 |
ENSMUST00000067582.7
|
Tmem62
|
transmembrane protein 62 |
| chr19_-_57197377 | 1.00 |
ENSMUST00000111546.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr15_+_77698889 | 1.00 |
ENSMUST00000096358.4
|
Apol7e
|
apolipoprotein L 7e |
| chr2_+_69135799 | 0.99 |
ENSMUST00000041865.7
|
Nostrin
|
nitric oxide synthase trafficker |
| chr15_+_88819584 | 0.99 |
ENSMUST00000024042.3
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr4_+_19575139 | 0.99 |
ENSMUST00000108253.1
ENSMUST00000029888.3 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr15_-_77447444 | 0.98 |
ENSMUST00000089469.5
|
Apol7b
|
apolipoprotein L 7b |
| chr17_-_70851189 | 0.98 |
ENSMUST00000059775.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_-_72795801 | 0.97 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr8_-_61902669 | 0.97 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr5_-_71548190 | 0.96 |
ENSMUST00000050129.5
|
Cox7b2
|
cytochrome c oxidase subunit VIIb2 |
| chr11_+_99864476 | 0.95 |
ENSMUST00000092694.3
|
Gm11559
|
predicted gene 11559 |
| chr2_+_162987330 | 0.94 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_-_95015416 | 0.94 |
ENSMUST00000132195.1
|
Zfp687
|
zinc finger protein 687 |
| chr12_+_14494561 | 0.94 |
ENSMUST00000052528.3
|
Gm9847
|
predicted pseudogene 9847 |
| chr3_+_85574109 | 0.93 |
ENSMUST00000127348.1
ENSMUST00000107672.1 ENSMUST00000107674.1 |
Pet112
|
PET112 homolog (S. cerevisiae) |
| chr8_+_22411340 | 0.93 |
ENSMUST00000033934.3
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr10_+_20148457 | 0.93 |
ENSMUST00000020173.8
|
Map7
|
microtubule-associated protein 7 |
| chr16_+_11322915 | 0.92 |
ENSMUST00000115814.3
|
Snx29
|
sorting nexin 29 |
| chr13_+_8885501 | 0.92 |
ENSMUST00000169314.2
|
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr9_-_103222063 | 0.92 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chr2_+_132846638 | 0.91 |
ENSMUST00000028835.6
ENSMUST00000110122.3 |
Crls1
|
cardiolipin synthase 1 |
| chr19_-_57197435 | 0.91 |
ENSMUST00000111550.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_+_44850393 | 0.90 |
ENSMUST00000136232.1
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr1_+_34851832 | 0.90 |
ENSMUST00000152654.1
|
Plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr4_-_141239453 | 0.90 |
ENSMUST00000168138.1
|
C630004L07Rik
|
RIKEN cDNA C630004L07 gene |
| chr7_+_125707893 | 0.90 |
ENSMUST00000069660.6
ENSMUST00000142464.1 |
D430042O09Rik
|
RIKEN cDNA D430042O09 gene |
| chr6_-_129717132 | 0.89 |
ENSMUST00000088046.1
|
Klri1
|
killer cell lectin-like receptor family I member 1 |
| chr10_-_18234930 | 0.89 |
ENSMUST00000052648.8
ENSMUST00000080860.6 ENSMUST00000173243.1 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr4_-_138863469 | 0.88 |
ENSMUST00000030524.7
ENSMUST00000102513.1 |
Pla2g5
|
phospholipase A2, group V |
| chrX_+_7722267 | 0.88 |
ENSMUST00000125991.1
ENSMUST00000148624.1 |
Wdr45
|
WD repeat domain 45 |
| chr5_-_107972864 | 0.88 |
ENSMUST00000153172.1
|
Fam69a
|
family with sequence similarity 69, member A |
| chr3_-_53863764 | 0.88 |
ENSMUST00000122330.1
ENSMUST00000146598.1 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr19_-_38125258 | 0.87 |
ENSMUST00000025951.6
|
Rbp4
|
retinol binding protein 4, plasma |
| chr8_-_38661508 | 0.86 |
ENSMUST00000118896.1
|
Sgcz
|
sarcoglycan zeta |
| chr4_-_147904680 | 0.85 |
ENSMUST00000105716.2
ENSMUST00000105715.1 ENSMUST00000105714.1 ENSMUST00000030884.3 |
Mfn2
|
mitofusin 2 |
| chr10_-_42478488 | 0.85 |
ENSMUST00000041024.8
|
Lace1
|
lactation elevated 1 |
| chr8_+_104961713 | 0.84 |
ENSMUST00000043183.7
|
Ces2g
|
carboxylesterase 2G |
| chr1_+_52845013 | 0.84 |
ENSMUST00000159352.1
ENSMUST00000044478.6 |
Hibch
|
3-hydroxyisobutyryl-Coenzyme A hydrolase |
| chr2_-_157571270 | 0.83 |
ENSMUST00000173378.1
|
Blcap
|
bladder cancer associated protein homolog (human) |
| chr14_-_30923547 | 0.83 |
ENSMUST00000170415.1
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr1_+_156558759 | 0.83 |
ENSMUST00000173929.1
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr19_+_55253369 | 0.83 |
ENSMUST00000043150.4
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr3_-_146812951 | 0.83 |
ENSMUST00000102515.3
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr5_-_138619702 | 0.82 |
ENSMUST00000063262.4
|
Zfp68
|
zinc finger protein 68 |
| chr5_-_52669677 | 0.82 |
ENSMUST00000031069.6
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meis2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.8 | 8.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.7 | 5.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.2 | 3.6 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.9 | 2.7 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.9 | 12.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.9 | 2.6 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.8 | 2.3 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.7 | 2.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.7 | 4.0 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.7 | 3.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.6 | 2.5 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.6 | 2.5 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.6 | 1.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 10.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.6 | 2.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 2.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.6 | 2.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 3.8 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.5 | 2.1 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.5 | 7.1 | GO:0006063 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.5 | 1.4 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.4 | 1.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 1.9 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 2.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.4 | 1.1 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 0.3 | 2.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.3 | 1.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.3 | 1.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 1.3 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.3 | 2.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.3 | 2.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 0.8 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 1.3 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 1.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 1.9 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 1.4 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 0.7 | GO:0060168 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.2 | 0.9 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.2 | 1.1 | GO:1903587 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 | 1.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 1.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.6 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 2.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 0.6 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 2.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 0.8 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 2.5 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.2 | 0.5 | GO:2000812 | response to rapamycin(GO:1901355) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 0.5 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 1.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.9 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 2.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.4 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.8 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 1.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 0.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.7 | GO:0023021 | sphingomyelin catabolic process(GO:0006685) termination of signal transduction(GO:0023021) |
| 0.1 | 0.4 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 0.7 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 1.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 6.0 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.6 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.6 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.5 | GO:0061295 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.3 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.9 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.9 | GO:1903874 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) ferrous iron transmembrane transport(GO:1903874) |
| 0.1 | 0.8 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 1.7 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 2.8 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.8 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.7 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 6.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.9 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.4 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.1 | 0.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.7 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 1.1 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 1.1 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.5 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.1 | 0.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 5.0 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 2.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 2.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 1.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) motor behavior(GO:0061744) |
| 0.1 | 1.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 2.7 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.5 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.9 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.6 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) zymogen granule exocytosis(GO:0070625) |
| 0.0 | 0.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.1 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 1.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.6 | GO:0002719 | negative regulation of cytokine production involved in immune response(GO:0002719) |
| 0.0 | 0.4 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 2.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 1.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 5.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.8 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 2.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 1.2 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 4.3 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 2.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.6 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 1.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.6 | GO:0030501 | positive regulation of bone mineralization(GO:0030501) |
| 0.0 | 2.8 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.9 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 10.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.8 | 8.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 2.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.5 | 2.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 2.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 2.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 5.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 2.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.2 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 1.9 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 20.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 1.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.0 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 4.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 4.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 1.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 5.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 7.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 4.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 2.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 20.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.2 | 1.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 1.0 | 6.9 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.9 | 2.7 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.8 | 12.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 3.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.6 | 2.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.6 | 1.8 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.6 | 1.7 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.5 | 19.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 2.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 1.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.5 | 1.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.4 | 4.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 2.1 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.4 | 1.7 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.4 | 5.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 1.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.4 | 2.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 1.9 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 1.1 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.3 | 1.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.7 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.3 | 2.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.3 | 2.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 0.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 1.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 3.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 1.8 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 2.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 0.9 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 0.7 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.2 | 0.6 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 2.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.6 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.2 | 5.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 0.5 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.2 | 0.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 0.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 3.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.7 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.9 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 1.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.7 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.3 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 1.4 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 3.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 1.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 1.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 2.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 6.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 1.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 2.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 5.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 2.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.9 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 4.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 5.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 5.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 8.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 1.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 4.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 4.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 6.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.9 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |