Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
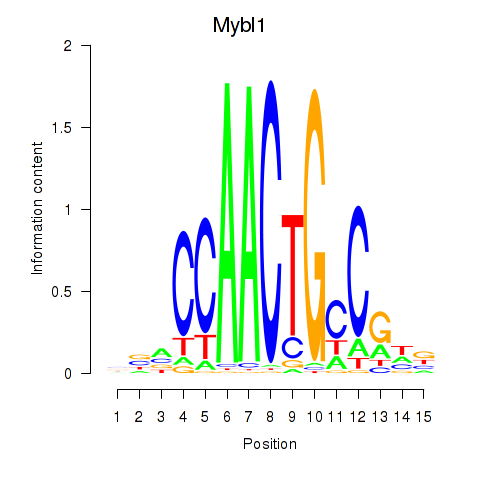
Results for Mybl1
Z-value: 2.00
Transcription factors associated with Mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl1
|
ENSMUSG00000025912.10 | myeloblastosis oncogene-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl1 | mm10_v2_chr1_-_9700209_9700329 | 0.46 | 5.0e-03 | Click! |
Activity profile of Mybl1 motif
Sorted Z-values of Mybl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_79886302 | 22.74 |
ENSMUST00000046091.5
|
Elane
|
elastase, neutrophil expressed |
| chr19_-_11640828 | 21.71 |
ENSMUST00000112984.2
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr14_-_56102458 | 17.85 |
ENSMUST00000015583.1
|
Ctsg
|
cathepsin G |
| chr11_+_87793722 | 17.12 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr11_+_87793470 | 16.17 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr11_+_58918004 | 15.52 |
ENSMUST00000108818.3
ENSMUST00000020792.5 |
Btnl10
|
butyrophilin-like 10 |
| chr4_-_119189949 | 15.20 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_46413486 | 13.87 |
ENSMUST00000071096.2
|
Hemgn
|
hemogen |
| chr11_+_58640394 | 13.51 |
ENSMUST00000075084.4
|
Trim58
|
tripartite motif-containing 58 |
| chr8_+_94172618 | 12.92 |
ENSMUST00000034214.6
|
Mt2
|
metallothionein 2 |
| chr3_-_106167564 | 12.39 |
ENSMUST00000063062.8
|
Chi3l3
|
chitinase 3-like 3 |
| chr2_+_84980458 | 10.17 |
ENSMUST00000028467.5
|
Prg2
|
proteoglycan 2, bone marrow |
| chr4_-_132533488 | 9.95 |
ENSMUST00000152993.1
ENSMUST00000067496.6 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr17_+_48247759 | 9.87 |
ENSMUST00000048065.5
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr19_-_17356631 | 9.61 |
ENSMUST00000174236.1
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr2_+_174450678 | 9.43 |
ENSMUST00000016399.5
|
Tubb1
|
tubulin, beta 1 class VI |
| chr15_-_96642311 | 9.43 |
ENSMUST00000088454.5
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr14_+_46760526 | 9.17 |
ENSMUST00000067426.4
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr12_+_109549157 | 9.10 |
ENSMUST00000128458.1
ENSMUST00000150851.1 |
Meg3
|
maternally expressed 3 |
| chr17_-_35085609 | 8.86 |
ENSMUST00000038507.6
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr15_-_103252810 | 8.56 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr2_-_150668198 | 8.20 |
ENSMUST00000028944.3
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr7_+_79660196 | 8.16 |
ENSMUST00000035977.7
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr3_-_69044697 | 8.06 |
ENSMUST00000136512.1
ENSMUST00000143454.1 ENSMUST00000107802.1 |
Trim59
|
tripartite motif-containing 59 |
| chr2_+_118814195 | 7.96 |
ENSMUST00000110842.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr6_+_124829540 | 7.85 |
ENSMUST00000150120.1
|
Cdca3
|
cell division cycle associated 3 |
| chr2_+_118814237 | 7.68 |
ENSMUST00000028803.7
ENSMUST00000126045.1 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr3_+_51661167 | 7.54 |
ENSMUST00000099106.3
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr14_+_65806066 | 7.49 |
ENSMUST00000139644.1
|
Pbk
|
PDZ binding kinase |
| chr2_+_118813995 | 7.47 |
ENSMUST00000134661.1
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chrX_-_102157065 | 7.43 |
ENSMUST00000056904.2
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr11_+_32276400 | 7.37 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr2_+_163054682 | 7.21 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr11_+_32296489 | 7.08 |
ENSMUST00000093207.3
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr6_+_124829582 | 7.08 |
ENSMUST00000024270.7
|
Cdca3
|
cell division cycle associated 3 |
| chr7_-_4397705 | 6.97 |
ENSMUST00000108590.2
|
Gp6
|
glycoprotein 6 (platelet) |
| chr3_+_30856014 | 6.94 |
ENSMUST00000108258.1
ENSMUST00000147697.1 |
Gpr160
|
G protein-coupled receptor 160 |
| chr11_+_5569679 | 6.93 |
ENSMUST00000109856.1
ENSMUST00000109855.1 ENSMUST00000118112.2 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr14_-_60086832 | 6.90 |
ENSMUST00000080368.5
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chrX_+_136270253 | 6.70 |
ENSMUST00000178632.1
ENSMUST00000053540.4 |
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr1_+_134182404 | 6.66 |
ENSMUST00000153856.1
ENSMUST00000082060.3 ENSMUST00000133701.1 ENSMUST00000132873.1 |
Chi3l1
|
chitinase 3-like 1 |
| chr1_+_134182150 | 6.58 |
ENSMUST00000156873.1
|
Chi3l1
|
chitinase 3-like 1 |
| chr4_-_118437331 | 6.51 |
ENSMUST00000006565.6
|
Cdc20
|
cell division cycle 20 |
| chr5_+_123749696 | 6.46 |
ENSMUST00000031366.7
|
Kntc1
|
kinetochore associated 1 |
| chr12_-_99883429 | 6.26 |
ENSMUST00000046485.3
|
Efcab11
|
EF-hand calcium binding domain 11 |
| chr11_-_72550255 | 6.17 |
ENSMUST00000021154.6
|
Spns3
|
spinster homolog 3 |
| chr10_+_75573448 | 5.98 |
ENSMUST00000006508.3
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr1_-_133753681 | 5.87 |
ENSMUST00000125659.1
ENSMUST00000165602.2 ENSMUST00000048953.7 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr4_-_117182623 | 5.74 |
ENSMUST00000065896.2
|
Kif2c
|
kinesin family member 2C |
| chr4_-_44167580 | 5.67 |
ENSMUST00000098098.2
|
Rnf38
|
ring finger protein 38 |
| chr1_+_153751946 | 5.65 |
ENSMUST00000183241.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr7_+_78913765 | 5.59 |
ENSMUST00000038142.8
|
Isg20
|
interferon-stimulated protein |
| chr17_-_26095487 | 5.57 |
ENSMUST00000025007.5
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr1_+_57995971 | 5.55 |
ENSMUST00000027202.8
|
Sgol2
|
shugoshin-like 2 (S. pombe) |
| chr7_-_127137807 | 5.37 |
ENSMUST00000049931.5
|
Spn
|
sialophorin |
| chr4_-_43499608 | 5.37 |
ENSMUST00000136005.1
ENSMUST00000054538.6 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr7_+_126862431 | 5.34 |
ENSMUST00000132808.1
|
Hirip3
|
HIRA interacting protein 3 |
| chr8_+_83955507 | 5.32 |
ENSMUST00000005607.8
|
Asf1b
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae) |
| chr2_-_170427828 | 5.27 |
ENSMUST00000013667.2
ENSMUST00000109152.2 ENSMUST00000068137.4 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr6_+_30723541 | 5.25 |
ENSMUST00000115127.1
|
Mest
|
mesoderm specific transcript |
| chr18_-_35649349 | 5.21 |
ENSMUST00000025211.4
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr3_+_30855946 | 5.20 |
ENSMUST00000108261.1
ENSMUST00000108259.1 ENSMUST00000166278.1 ENSMUST00000046748.6 |
Gpr160
|
G protein-coupled receptor 160 |
| chr10_+_79930419 | 5.19 |
ENSMUST00000131118.1
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr17_-_23645264 | 5.09 |
ENSMUST00000024696.7
|
Mmp25
|
matrix metallopeptidase 25 |
| chr12_+_69168808 | 5.09 |
ENSMUST00000110621.1
|
Lrr1
|
leucine rich repeat protein 1 |
| chr2_+_122637844 | 5.07 |
ENSMUST00000047498.8
|
AA467197
|
expressed sequence AA467197 |
| chrX_+_50841434 | 5.02 |
ENSMUST00000114887.2
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene |
| chr2_-_35979624 | 5.00 |
ENSMUST00000028248.4
ENSMUST00000112976.2 |
Ttll11
|
tubulin tyrosine ligase-like family, member 11 |
| chr15_+_79891631 | 4.99 |
ENSMUST00000177350.1
ENSMUST00000177483.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr9_+_27030159 | 4.94 |
ENSMUST00000073127.7
ENSMUST00000086198.4 |
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr3_-_92485886 | 4.87 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr13_+_108316395 | 4.81 |
ENSMUST00000171178.1
|
Depdc1b
|
DEP domain containing 1B |
| chr15_-_74983430 | 4.77 |
ENSMUST00000023250.4
ENSMUST00000166694.1 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chrX_+_69360294 | 4.72 |
ENSMUST00000033532.6
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr11_-_69605829 | 4.71 |
ENSMUST00000047889.6
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr9_-_39603635 | 4.57 |
ENSMUST00000119722.1
|
AW551984
|
expressed sequence AW551984 |
| chr6_-_128891105 | 4.57 |
ENSMUST00000178918.1
ENSMUST00000160290.1 |
BC035044
|
cDNA sequence BC035044 |
| chr11_-_33513736 | 4.46 |
ENSMUST00000102815.3
|
Ranbp17
|
RAN binding protein 17 |
| chr3_+_10012548 | 4.41 |
ENSMUST00000029046.8
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr14_-_31577318 | 4.38 |
ENSMUST00000112027.2
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr19_+_60755947 | 4.33 |
ENSMUST00000088237.4
|
Nanos1
|
nanos homolog 1 (Drosophila) |
| chr1_-_193370260 | 4.26 |
ENSMUST00000016323.4
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr2_+_29124106 | 4.24 |
ENSMUST00000129544.1
|
Setx
|
senataxin |
| chr9_+_64811313 | 4.22 |
ENSMUST00000038890.5
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr11_-_71033462 | 4.21 |
ENSMUST00000156068.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr16_+_93883895 | 4.20 |
ENSMUST00000023666.4
ENSMUST00000117099.1 ENSMUST00000142316.1 |
Chaf1b
|
chromatin assembly factor 1, subunit B (p60) |
| chr14_+_65805832 | 4.18 |
ENSMUST00000022612.3
|
Pbk
|
PDZ binding kinase |
| chr10_+_79664553 | 4.04 |
ENSMUST00000020554.6
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr8_+_83608175 | 4.02 |
ENSMUST00000005620.8
|
Dnajb1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr13_-_23762378 | 4.00 |
ENSMUST00000091701.2
|
Hist1h3a
|
histone cluster 1, H3a |
| chr9_+_96258697 | 3.99 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chr7_+_99594605 | 3.95 |
ENSMUST00000162290.1
|
Arrb1
|
arrestin, beta 1 |
| chr13_-_49652714 | 3.94 |
ENSMUST00000021818.7
|
Cenpp
|
centromere protein P |
| chr1_-_94052553 | 3.92 |
ENSMUST00000027507.6
|
Pdcd1
|
programmed cell death 1 |
| chr19_-_41802028 | 3.90 |
ENSMUST00000026150.8
ENSMUST00000177495.1 ENSMUST00000163265.1 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr7_+_100285478 | 3.85 |
ENSMUST00000057023.5
|
P4ha3
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide III |
| chr7_+_43437073 | 3.85 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr4_-_44167988 | 3.82 |
ENSMUST00000143337.1
|
Rnf38
|
ring finger protein 38 |
| chr2_+_172393900 | 3.81 |
ENSMUST00000109136.2
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr2_+_71117923 | 3.79 |
ENSMUST00000028403.2
|
Cybrd1
|
cytochrome b reductase 1 |
| chr11_+_58379036 | 3.79 |
ENSMUST00000013787.4
ENSMUST00000108826.2 |
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr4_-_44167905 | 3.70 |
ENSMUST00000102934.2
|
Rnf38
|
ring finger protein 38 |
| chr4_+_154869585 | 3.64 |
ENSMUST00000079269.7
ENSMUST00000163732.1 ENSMUST00000080559.6 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr12_-_69790660 | 3.60 |
ENSMUST00000021377.4
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr6_-_57825055 | 3.59 |
ENSMUST00000127485.1
|
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr18_+_34624621 | 3.58 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr8_-_25091341 | 3.57 |
ENSMUST00000125466.1
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr12_+_111406809 | 3.56 |
ENSMUST00000150384.1
|
A230065H16Rik
|
RIKEN cDNA A230065H16 gene |
| chr2_-_164356507 | 3.53 |
ENSMUST00000109367.3
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr5_-_100562836 | 3.52 |
ENSMUST00000097437.2
|
Plac8
|
placenta-specific 8 |
| chr8_+_83715504 | 3.51 |
ENSMUST00000109810.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr1_-_193370225 | 3.48 |
ENSMUST00000169907.1
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr10_-_79874233 | 3.47 |
ENSMUST00000166023.1
ENSMUST00000167707.1 ENSMUST00000165601.1 |
BC005764
|
cDNA sequence BC005764 |
| chr17_-_46629420 | 3.40 |
ENSMUST00000044442.8
|
Ptk7
|
PTK7 protein tyrosine kinase 7 |
| chr11_+_101325063 | 3.38 |
ENSMUST00000041095.7
ENSMUST00000107264.1 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr8_+_86745679 | 3.38 |
ENSMUST00000098532.2
|
Gm10638
|
predicted gene 10638 |
| chr19_+_4154606 | 3.35 |
ENSMUST00000061086.8
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr17_-_79355082 | 3.34 |
ENSMUST00000068958.7
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr6_-_57825144 | 3.31 |
ENSMUST00000114297.2
|
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr1_-_191575534 | 3.30 |
ENSMUST00000027933.5
|
Dtl
|
denticleless homolog (Drosophila) |
| chr18_-_73815392 | 3.30 |
ENSMUST00000025439.3
|
Me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr5_-_114813943 | 3.26 |
ENSMUST00000061251.5
ENSMUST00000112160.3 |
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chr9_-_95858320 | 3.22 |
ENSMUST00000178038.1
|
1700065D16Rik
|
RIKEN cDNA 1700065D16 gene |
| chr1_-_169531447 | 3.21 |
ENSMUST00000111368.1
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr15_-_96642883 | 3.19 |
ENSMUST00000088452.4
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr19_-_5894100 | 3.17 |
ENSMUST00000055911.4
|
Tigd3
|
tigger transposable element derived 3 |
| chr6_-_143100028 | 3.16 |
ENSMUST00000111758.2
ENSMUST00000171349.1 ENSMUST00000087485.4 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr2_+_109280738 | 3.07 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr15_-_78572754 | 3.07 |
ENSMUST00000043214.6
|
Rac2
|
RAS-related C3 botulinum substrate 2 |
| chr11_-_33513626 | 3.06 |
ENSMUST00000037522.7
|
Ranbp17
|
RAN binding protein 17 |
| chr11_-_120348475 | 3.06 |
ENSMUST00000062147.7
ENSMUST00000128055.1 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr12_+_111971545 | 3.04 |
ENSMUST00000079009.5
|
Tdrd9
|
tudor domain containing 9 |
| chr19_-_11623643 | 3.03 |
ENSMUST00000164792.1
ENSMUST00000025583.5 |
Ms4a2
|
membrane-spanning 4-domains, subfamily A, member 2 |
| chr4_+_100776664 | 3.02 |
ENSMUST00000030257.8
ENSMUST00000097955.2 |
Cachd1
|
cache domain containing 1 |
| chr2_+_121506715 | 3.01 |
ENSMUST00000028676.5
|
Wdr76
|
WD repeat domain 76 |
| chr2_+_156840966 | 3.01 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr1_+_180935022 | 2.97 |
ENSMUST00000037361.8
|
Lefty1
|
left right determination factor 1 |
| chr16_+_10170228 | 2.97 |
ENSMUST00000044103.5
|
Rpl39l
|
ribosomal protein L39-like |
| chr2_+_119112793 | 2.96 |
ENSMUST00000140939.1
ENSMUST00000028795.3 |
Rad51
|
RAD51 homolog |
| chr11_+_54304005 | 2.96 |
ENSMUST00000000145.5
ENSMUST00000138515.1 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr4_+_45184815 | 2.87 |
ENSMUST00000134280.1
ENSMUST00000044773.5 |
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr8_+_83715177 | 2.86 |
ENSMUST00000019576.8
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr4_-_43045686 | 2.85 |
ENSMUST00000107956.1
ENSMUST00000107957.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr8_-_105851981 | 2.84 |
ENSMUST00000040776.4
|
Cenpt
|
centromere protein T |
| chr11_-_109472611 | 2.84 |
ENSMUST00000168740.1
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr18_+_67800101 | 2.82 |
ENSMUST00000025425.5
|
Cep192
|
centrosomal protein 192 |
| chr6_-_83536215 | 2.82 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr19_-_42086338 | 2.81 |
ENSMUST00000051772.8
|
Morn4
|
MORN repeat containing 4 |
| chr11_+_95337012 | 2.80 |
ENSMUST00000037502.6
|
Fam117a
|
family with sequence similarity 117, member A |
| chr11_+_98551097 | 2.79 |
ENSMUST00000081033.6
ENSMUST00000107511.1 ENSMUST00000107509.1 ENSMUST00000017339.5 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr15_-_73184840 | 2.79 |
ENSMUST00000044113.10
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr13_-_117025505 | 2.77 |
ENSMUST00000022239.6
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr8_+_83715239 | 2.75 |
ENSMUST00000172396.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr11_+_82892116 | 2.73 |
ENSMUST00000018988.5
|
Fndc8
|
fibronectin type III domain containing 8 |
| chrX_+_136245065 | 2.73 |
ENSMUST00000048687.4
|
Wbp5
|
WW domain binding protein 5 |
| chr5_+_123076275 | 2.66 |
ENSMUST00000067505.8
ENSMUST00000111619.3 ENSMUST00000160344.1 |
Tmem120b
|
transmembrane protein 120B |
| chr13_-_59769751 | 2.66 |
ENSMUST00000057115.6
|
Isca1
|
iron-sulfur cluster assembly 1 homolog (S. cerevisiae) |
| chr6_-_40999479 | 2.66 |
ENSMUST00000166306.1
|
Gm2663
|
predicted gene 2663 |
| chr1_-_167285110 | 2.65 |
ENSMUST00000027839.8
|
Uck2
|
uridine-cytidine kinase 2 |
| chr7_-_141279121 | 2.64 |
ENSMUST00000167790.1
ENSMUST00000046156.6 |
Sct
|
secretin |
| chr15_-_99251929 | 2.62 |
ENSMUST00000041190.9
ENSMUST00000163506.1 |
Mcrs1
|
microspherule protein 1 |
| chrX_+_36743659 | 2.60 |
ENSMUST00000047655.6
|
Slc25a43
|
solute carrier family 25, member 43 |
| chr2_+_31950257 | 2.60 |
ENSMUST00000001920.7
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr8_-_68121527 | 2.59 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chr15_-_89425795 | 2.58 |
ENSMUST00000168376.1
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr1_-_33669745 | 2.57 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr3_-_95995835 | 2.56 |
ENSMUST00000143485.1
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr11_+_68901538 | 2.56 |
ENSMUST00000073471.6
ENSMUST00000101014.2 ENSMUST00000128952.1 ENSMUST00000167436.1 |
Rpl26
|
ribosomal protein L26 |
| chr4_+_103619580 | 2.54 |
ENSMUST00000106827.1
|
Dab1
|
disabled 1 |
| chr12_-_114416895 | 2.53 |
ENSMUST00000179796.1
|
Ighv6-5
|
immunoglobulin heavy variable V6-5 |
| chr1_-_174250976 | 2.53 |
ENSMUST00000061990.4
|
Olfr419
|
olfactory receptor 419 |
| chr10_+_14523062 | 2.51 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr4_-_44168339 | 2.50 |
ENSMUST00000045793.8
|
Rnf38
|
ring finger protein 38 |
| chr4_-_129558387 | 2.49 |
ENSMUST00000067240.4
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr7_-_35537677 | 2.48 |
ENSMUST00000127472.1
ENSMUST00000032701.7 |
Tdrd12
|
tudor domain containing 12 |
| chr7_+_126861947 | 2.48 |
ENSMUST00000037248.3
|
Hirip3
|
HIRA interacting protein 3 |
| chr8_-_71272367 | 2.43 |
ENSMUST00000110071.2
|
Haus8
|
4HAUS augmin-like complex, subunit 8 |
| chr9_+_57827284 | 2.43 |
ENSMUST00000163186.1
|
Gm17231
|
predicted gene 17231 |
| chr14_-_65425453 | 2.42 |
ENSMUST00000059339.5
|
Pnoc
|
prepronociceptin |
| chr13_+_24614608 | 2.41 |
ENSMUST00000091694.3
|
Fam65b
|
family with sequence similarity 65, member B |
| chr1_+_40324570 | 2.40 |
ENSMUST00000095020.3
|
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr4_-_131967824 | 2.39 |
ENSMUST00000146443.1
ENSMUST00000135579.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr1_+_153425162 | 2.39 |
ENSMUST00000042373.5
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr8_+_104250869 | 2.38 |
ENSMUST00000034342.5
|
Cklf
|
chemokine-like factor |
| chr3_+_153973436 | 2.38 |
ENSMUST00000089948.5
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr5_+_110286306 | 2.36 |
ENSMUST00000007296.5
ENSMUST00000112482.1 |
Pole
|
polymerase (DNA directed), epsilon |
| chr2_+_91650169 | 2.35 |
ENSMUST00000090614.4
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr11_-_109473220 | 2.35 |
ENSMUST00000070872.6
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr9_-_15045378 | 2.32 |
ENSMUST00000164273.1
|
Panx1
|
pannexin 1 |
| chrX_+_135839034 | 2.32 |
ENSMUST00000173804.1
ENSMUST00000113136.1 |
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chr17_+_8849974 | 2.31 |
ENSMUST00000115720.1
|
Pde10a
|
phosphodiesterase 10A |
| chr2_+_121506748 | 2.31 |
ENSMUST00000099473.3
ENSMUST00000110602.2 |
Wdr76
|
WD repeat domain 76 |
| chr11_-_43601540 | 2.28 |
ENSMUST00000020672.4
|
Fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr4_-_129558355 | 2.27 |
ENSMUST00000167288.1
ENSMUST00000134336.1 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr16_+_10170216 | 2.27 |
ENSMUST00000121292.1
|
Rpl39l
|
ribosomal protein L39-like |
| chr5_+_110514885 | 2.27 |
ENSMUST00000141532.1
|
A630023P12Rik
|
RIKEN cDNA A630023P12 gene |
| chr7_-_4812351 | 2.21 |
ENSMUST00000079496.7
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr18_-_62179948 | 2.21 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr8_-_24948771 | 2.16 |
ENSMUST00000119720.1
ENSMUST00000121438.2 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr17_+_29093763 | 2.16 |
ENSMUST00000023829.6
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.1 | 33.3 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 7.6 | 22.7 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) neutrophil mediated killing of fungus(GO:0070947) |
| 3.9 | 27.0 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 3.5 | 10.5 | GO:0002215 | defense response to nematode(GO:0002215) |
| 2.2 | 6.5 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 2.1 | 12.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 2.0 | 10.0 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 2.0 | 5.9 | GO:0045763 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.8 | 12.9 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.8 | 5.4 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 1.6 | 6.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.4 | 4.2 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 1.4 | 6.9 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 1.4 | 8.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 1.3 | 3.9 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 1.3 | 23.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.2 | 3.6 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 1.2 | 4.7 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.2 | 3.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.1 | 5.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.1 | 5.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.1 | 8.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.0 | 3.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.0 | 8.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 1.0 | 9.1 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 1.0 | 2.9 | GO:0043465 | negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.9 | 4.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.9 | 6.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.9 | 2.6 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of somatostatin secretion(GO:0090274) |
| 0.9 | 2.6 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.8 | 2.5 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.8 | 2.5 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.8 | 11.6 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.8 | 4.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.8 | 2.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.8 | 3.0 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.7 | 6.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.7 | 3.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.7 | 3.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.7 | 4.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.7 | 8.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.7 | 4.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.7 | 4.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.7 | 3.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 | 1.9 | GO:1902174 | striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.6 | 2.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.6 | 5.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.6 | 1.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.6 | 5.0 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.6 | 1.9 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.6 | 4.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 2.9 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.6 | 0.6 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.5 | 1.6 | GO:1904100 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.5 | 2.1 | GO:2000705 | negative regulation of anion channel activity(GO:0010360) regulation of dense core granule biogenesis(GO:2000705) |
| 0.5 | 10.4 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.5 | 5.6 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.5 | 2.0 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.5 | 7.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.5 | 1.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.5 | 2.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 1.4 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.5 | 1.9 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.5 | 8.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.5 | 4.8 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.5 | 3.3 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.4 | 14.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.7 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.4 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 2.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 1.2 | GO:0019661 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 1.6 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.4 | 1.2 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.4 | 2.4 | GO:0032439 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.4 | 3.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.4 | 1.1 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.4 | 2.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 1.7 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.3 | 1.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 2.6 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 6.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 4.6 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.3 | 9.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 3.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 3.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 1.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 1.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 2.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 0.6 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.3 | 0.9 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 0.9 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 1.5 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.3 | 0.9 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.3 | 1.5 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.3 | 2.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 1.5 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.3 | 1.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.2 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.3 | 1.7 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.3 | 0.9 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 3.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 1.7 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 1.4 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.3 | 4.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.3 | 1.0 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 | 3.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.3 | 0.8 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.3 | 3.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.3 | 2.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 1.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 3.7 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 1.2 | GO:0032901 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) regulation of neurotrophin production(GO:0032899) positive regulation of neurotrophin production(GO:0032901) |
| 0.2 | 2.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 0.7 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 2.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 0.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 4.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 15.0 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.2 | 1.6 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.2 | 1.4 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 2.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 1.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 1.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 4.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.2 | 1.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.2 | 18.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 1.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.8 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.2 | 0.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 3.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 3.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 2.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 2.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 4.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 2.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 1.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.2 | 0.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 0.7 | GO:0035087 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.2 | 3.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.5 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 2.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 1.6 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.2 | 1.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 2.8 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 0.9 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.2 | 0.5 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.2 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 1.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 0.3 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) |
| 0.2 | 1.2 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 3.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 1.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 4.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 6.5 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 2.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 4.0 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 4.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.5 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 0.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.1 | 0.9 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.5 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 0.3 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.9 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 2.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.0 | GO:0098909 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 5.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.7 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 | 6.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 1.1 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.6 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.5 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 0.4 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 2.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.6 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.3 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.3 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 1.4 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 1.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 1.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 1.9 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 1.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 1.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.8 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.9 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.1 | 1.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.3 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.1 | 3.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 0.8 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 9.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.3 | GO:0000237 | leptotene(GO:0000237) |
| 0.1 | 3.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 5.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.3 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 1.3 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 3.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 3.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 2.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.7 | GO:0070262 | response to morphine(GO:0043278) peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 2.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 5.1 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 1.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 3.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.9 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 1.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.9 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 2.7 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 1.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.5 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.4 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 1.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.9 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 3.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 3.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 2.0 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 13.6 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 1.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 1.4 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.7 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 1.7 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.7 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.9 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.8 | GO:2001252 | positive regulation of chromosome organization(GO:2001252) |
| 0.0 | 0.3 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.0 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.7 | GO:0001649 | osteoblast differentiation(GO:0001649) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.1 | 33.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.8 | 14.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.6 | 4.9 | GO:0042585 | nuclear condensin complex(GO:0000799) germinal vesicle(GO:0042585) |
| 1.4 | 7.2 | GO:0031523 | Myb complex(GO:0031523) |
| 1.4 | 4.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.0 | 4.0 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.8 | 28.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.7 | 1.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.7 | 2.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.6 | 7.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.6 | 3.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.6 | 1.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.6 | 3.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.6 | 6.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.5 | 3.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 2.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.5 | 4.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 2.5 | GO:1990923 | PET complex(GO:1990923) |
| 0.5 | 2.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 3.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 4.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 2.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.4 | 2.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 2.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 1.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.4 | 2.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.4 | 2.6 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 8.7 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.3 | 5.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 3.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 1.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 0.9 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 7.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 2.8 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 2.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 0.8 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.3 | 1.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 1.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 5.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 1.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 4.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 5.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 1.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 1.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 3.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 0.9 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 1.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 1.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 14.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.7 | GO:0031011 | Ino80 complex(GO:0031011) INO80-type complex(GO:0097346) |
| 0.2 | 0.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 0.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 7.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 0.8 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 2.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 22.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 1.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 3.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 1.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 4.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 21.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.2 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 7.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 7.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 5.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 2.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 9.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 8.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 4.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 7.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 3.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 5.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 9.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 2.9 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 8.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 4.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 17.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 14.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 1.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 9.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 3.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 6.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 2.7 | 8.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 2.5 | 12.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 2.0 | 10.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.9 | 7.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.8 | 7.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 1.5 | 6.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.4 | 4.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 1.4 | 5.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.3 | 4.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.2 | 5.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.1 | 3.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.1 | 3.4 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 1.1 | 3.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.1 | 9.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.1 | 8.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.1 | 6.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.1 | 3.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 1.0 | 4.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.0 | 4.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 1.0 | 4.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.9 | 9.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.9 | 3.5 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.8 | 4.0 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.8 | 2.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.7 | 3.0 | GO:0038100 | nodal binding(GO:0038100) |
| 0.7 | 8.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.7 | 2.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.7 | 7.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.7 | 28.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.7 | 2.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.7 | 8.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.6 | 1.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 33.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.6 | 5.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 2.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.5 | 2.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.5 | 8.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 2.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 2.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.5 | 6.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.5 | 3.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 1.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.4 | 3.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 1.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 1.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 2.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 5.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.4 | 4.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.4 | 2.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 1.1 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.4 | 2.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 2.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 2.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 2.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.3 | 2.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 2.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 0.9 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 1.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.3 | 0.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.3 | 1.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 1.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 4.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 9.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 0.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.2 | 1.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 3.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.9 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 4.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 1.6 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 4.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 11.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 1.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.2 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 3.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 3.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 3.8 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 5.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 1.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 6.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 22.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 3.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 5.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 1.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.7 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 21.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 4.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 11.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.6 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 2.7 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 2.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 5.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 4.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 5.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 2.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 3.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.7 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.8 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 3.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 8.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 16.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 7.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 4.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 3.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 4.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 1.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 2.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 2.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 13.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0046625 | thioesterase binding(GO:0031996) sphingolipid binding(GO:0046625) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 1.1 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 33.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.6 | 30.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.5 | 5.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 10.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 19.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 3.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.3 | 13.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 4.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 4.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 10.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 5.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 10.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 5.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 4.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.9 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 4.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 4.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 5.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 6.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 43.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.4 | 8.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 8.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 14.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 8.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.4 | 6.7 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.4 | 39.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 1.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 2.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 7.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 4.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 10.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 5.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 3.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 9.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 3.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 11.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 2.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 4.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 3.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 6.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 5.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 4.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 10.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 3.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 8.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 4.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 6.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 12.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 4.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 2.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 4.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 3.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 9.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |