Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
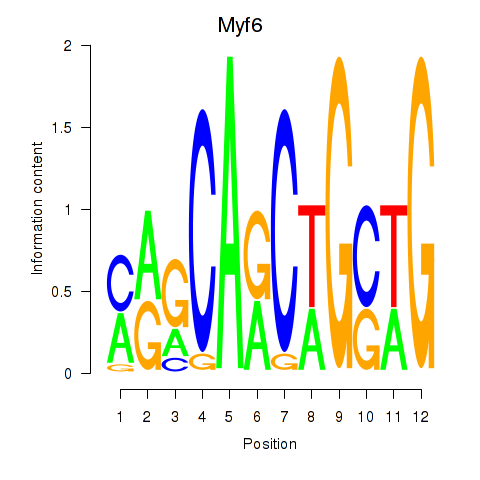
Results for Myf6
Z-value: 2.25
Transcription factors associated with Myf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myf6
|
ENSMUSG00000035923.3 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myf6 | mm10_v2_chr10_-_107494719_107494742 | -0.39 | 1.8e-02 | Click! |
Activity profile of Myf6 motif
Sorted Z-values of Myf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_62087261 | 23.15 |
ENSMUST00000107488.3
ENSMUST00000107472.1 ENSMUST00000084531.4 |
Mup3
|
major urinary protein 3 |
| chr4_-_60070411 | 21.57 |
ENSMUST00000079697.3
ENSMUST00000125282.1 ENSMUST00000166098.1 |
Mup7
|
major urinary protein 7 |
| chr4_-_60501903 | 21.25 |
ENSMUST00000084548.4
ENSMUST00000103012.3 ENSMUST00000107499.3 |
Mup1
|
major urinary protein 1 |
| chr4_-_60662358 | 21.17 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr4_-_61674094 | 21.13 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_60421933 | 20.71 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_61303998 | 20.43 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_60741275 | 19.90 |
ENSMUST00000117932.1
|
Mup12
|
major urinary protein 12 |
| chr4_-_60222580 | 19.64 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_61303802 | 19.56 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr10_+_128971191 | 19.54 |
ENSMUST00000181142.1
|
9030616G12Rik
|
RIKEN cDNA 9030616G12 gene |
| chr10_+_87860030 | 19.51 |
ENSMUST00000062862.6
|
Igf1
|
insulin-like growth factor 1 |
| chr4_-_60139857 | 18.00 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr10_-_128960965 | 14.03 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr4_-_60582152 | 12.14 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr6_-_85707858 | 11.29 |
ENSMUST00000179613.1
|
Gm4477
|
predicted gene 4477 |
| chr16_+_91269759 | 11.02 |
ENSMUST00000056882.5
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr15_-_86033777 | 9.71 |
ENSMUST00000016172.7
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) |
| chr7_-_114562945 | 9.31 |
ENSMUST00000119712.1
ENSMUST00000032908.8 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr4_+_60003438 | 9.12 |
ENSMUST00000107517.1
ENSMUST00000107520.1 |
Mup6
|
major urinary protein 6 |
| chr1_+_167598450 | 9.09 |
ENSMUST00000111386.1
ENSMUST00000111384.1 |
Rxrg
|
retinoid X receptor gamma |
| chr12_+_108334341 | 8.61 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr1_+_167598384 | 8.43 |
ENSMUST00000015987.3
|
Rxrg
|
retinoid X receptor gamma |
| chr16_-_46496772 | 8.35 |
ENSMUST00000149901.1
ENSMUST00000096052.2 |
Pvrl3
|
poliovirus receptor-related 3 |
| chr11_+_99879187 | 8.28 |
ENSMUST00000078442.3
|
Gm11567
|
predicted gene 11567 |
| chr15_-_3582596 | 8.19 |
ENSMUST00000161770.1
|
Ghr
|
growth hormone receptor |
| chr10_+_127866457 | 8.06 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr7_+_51879041 | 7.98 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chr1_+_72824482 | 7.79 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr11_+_99864476 | 7.73 |
ENSMUST00000092694.3
|
Gm11559
|
predicted gene 11559 |
| chr16_-_18089022 | 7.38 |
ENSMUST00000132241.1
ENSMUST00000139861.1 ENSMUST00000003620.5 |
Prodh
|
proline dehydrogenase |
| chr18_+_60803838 | 7.00 |
ENSMUST00000050487.8
ENSMUST00000097563.2 ENSMUST00000167610.1 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr7_+_51878967 | 6.96 |
ENSMUST00000051912.6
|
Gas2
|
growth arrest specific 2 |
| chr15_-_89373810 | 6.83 |
ENSMUST00000167643.2
|
Sco2
|
SCO cytochrome oxidase deficient homolog 2 (yeast) |
| chr1_-_121327672 | 6.77 |
ENSMUST00000159085.1
ENSMUST00000159125.1 ENSMUST00000161818.1 |
Insig2
|
insulin induced gene 2 |
| chr14_+_65968483 | 6.71 |
ENSMUST00000022616.6
|
Clu
|
clusterin |
| chr2_+_34772089 | 6.59 |
ENSMUST00000028222.6
ENSMUST00000100171.2 |
Hspa5
|
heat shock protein 5 |
| chr17_-_34000257 | 6.44 |
ENSMUST00000087189.6
ENSMUST00000173075.1 ENSMUST00000172760.1 ENSMUST00000172912.1 ENSMUST00000025181.10 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr1_-_121327734 | 6.36 |
ENSMUST00000160968.1
ENSMUST00000162582.1 |
Insig2
|
insulin induced gene 2 |
| chr2_-_24048857 | 6.35 |
ENSMUST00000114497.1
|
Hnmt
|
histamine N-methyltransferase |
| chr10_+_116301374 | 6.32 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr1_-_121328024 | 6.26 |
ENSMUST00000003818.7
|
Insig2
|
insulin induced gene 2 |
| chr9_-_43239816 | 6.18 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr14_-_79662084 | 6.09 |
ENSMUST00000165835.1
|
Lect1
|
leukocyte cell derived chemotaxin 1 |
| chr5_+_30588078 | 6.04 |
ENSMUST00000066295.2
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr7_-_142229971 | 6.02 |
ENSMUST00000097942.2
|
Krtap5-5
|
keratin associated protein 5-5 |
| chr16_-_46496955 | 6.02 |
ENSMUST00000023335.6
ENSMUST00000023334.8 |
Pvrl3
|
poliovirus receptor-related 3 |
| chr1_+_57377593 | 5.94 |
ENSMUST00000042734.2
|
1700066M21Rik
|
RIKEN cDNA 1700066M21 gene |
| chr3_+_94372794 | 5.83 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr1_-_121327776 | 5.79 |
ENSMUST00000160688.1
|
Insig2
|
insulin induced gene 2 |
| chr15_-_39857459 | 5.70 |
ENSMUST00000022915.3
ENSMUST00000110306.1 |
Dpys
|
dihydropyrimidinase |
| chr3_-_89322883 | 5.61 |
ENSMUST00000029673.5
|
Efna3
|
ephrin A3 |
| chr2_-_181043540 | 5.50 |
ENSMUST00000124400.1
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr19_+_26605106 | 5.44 |
ENSMUST00000025862.7
ENSMUST00000176030.1 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_84011442 | 5.37 |
ENSMUST00000056686.5
|
2210011C24Rik
|
RIKEN cDNA 2210011C24 gene |
| chr14_-_79662148 | 5.35 |
ENSMUST00000022603.7
|
Lect1
|
leukocyte cell derived chemotaxin 1 |
| chr7_-_99626936 | 5.30 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chr5_-_53213447 | 5.27 |
ENSMUST00000031090.6
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr17_+_35320529 | 5.24 |
ENSMUST00000105041.3
ENSMUST00000073208.5 |
H2-Q1
|
histocompatibility 2, Q region locus 1 |
| chr17_+_4994904 | 5.20 |
ENSMUST00000092723.4
ENSMUST00000115797.2 |
Arid1b
|
AT rich interactive domain 1B (SWI-like) |
| chr7_+_44590886 | 5.20 |
ENSMUST00000107906.3
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr1_-_180195981 | 5.20 |
ENSMUST00000027766.6
ENSMUST00000161814.1 |
Adck3
|
aarF domain containing kinase 3 |
| chr1_-_136260873 | 5.15 |
ENSMUST00000086395.5
|
Gpr25
|
G protein-coupled receptor 25 |
| chr11_+_104231390 | 5.15 |
ENSMUST00000106992.3
|
Mapt
|
microtubule-associated protein tau |
| chr10_-_20548361 | 5.07 |
ENSMUST00000164195.1
|
Pde7b
|
phosphodiesterase 7B |
| chr6_-_120294559 | 5.07 |
ENSMUST00000057283.7
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr5_+_43233928 | 5.06 |
ENSMUST00000114066.1
ENSMUST00000114065.1 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chrX_-_72656135 | 5.05 |
ENSMUST00000055966.6
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr17_-_46752170 | 5.05 |
ENSMUST00000121671.1
ENSMUST00000059844.6 |
Cnpy3
|
canopy 3 homolog (zebrafish) |
| chr14_+_65971164 | 4.96 |
ENSMUST00000144619.1
|
Clu
|
clusterin |
| chr18_-_61536522 | 4.90 |
ENSMUST00000171629.1
|
Arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr8_-_94696223 | 4.83 |
ENSMUST00000034227.4
|
Pllp
|
plasma membrane proteolipid |
| chr9_+_55326913 | 4.80 |
ENSMUST00000085754.3
ENSMUST00000034862.4 |
AI118078
|
expressed sequence AI118078 |
| chr5_+_43233463 | 4.80 |
ENSMUST00000169035.1
ENSMUST00000166713.1 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr11_+_115163333 | 4.76 |
ENSMUST00000021077.3
|
Slc9a3r1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
| chr4_+_133553370 | 4.75 |
ENSMUST00000042706.2
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr19_-_42202150 | 4.67 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr15_+_88819584 | 4.49 |
ENSMUST00000024042.3
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr17_-_57228003 | 4.48 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr19_+_7056731 | 4.48 |
ENSMUST00000040261.5
|
Macrod1
|
MACRO domain containing 1 |
| chr10_-_20548320 | 4.47 |
ENSMUST00000169404.1
|
Pde7b
|
phosphodiesterase 7B |
| chr1_-_133921393 | 4.44 |
ENSMUST00000048432.5
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr5_-_87591582 | 4.41 |
ENSMUST00000031201.7
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr7_+_24481991 | 4.40 |
ENSMUST00000068023.7
|
Cadm4
|
cell adhesion molecule 4 |
| chr11_+_99785191 | 4.36 |
ENSMUST00000105059.2
|
Krtap4-9
|
keratin associated protein 4-9 |
| chr10_+_127776374 | 4.33 |
ENSMUST00000136223.1
ENSMUST00000052652.6 |
Rdh9
|
retinol dehydrogenase 9 |
| chr4_-_66404512 | 4.32 |
ENSMUST00000068214.4
|
Astn2
|
astrotactin 2 |
| chr11_+_99873389 | 4.26 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr17_-_34804546 | 4.12 |
ENSMUST00000025223.8
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr17_-_87282793 | 4.06 |
ENSMUST00000146560.2
|
4833418N02Rik
|
RIKEN cDNA 4833418N02 gene |
| chr7_-_12998140 | 3.96 |
ENSMUST00000032539.7
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr10_-_114801364 | 3.87 |
ENSMUST00000061632.7
|
Trhde
|
TRH-degrading enzyme |
| chr15_+_76268076 | 3.85 |
ENSMUST00000074173.3
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr10_+_7589885 | 3.85 |
ENSMUST00000130590.1
ENSMUST00000135907.1 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr2_-_26604267 | 3.80 |
ENSMUST00000028286.5
|
Agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr11_-_99620432 | 3.71 |
ENSMUST00000073853.2
|
Gm11562
|
predicted gene 11562 |
| chr10_-_81545175 | 3.69 |
ENSMUST00000043604.5
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr13_+_93771656 | 3.64 |
ENSMUST00000091403.4
|
Arsb
|
arylsulfatase B |
| chr9_-_114844090 | 3.64 |
ENSMUST00000047013.3
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr4_+_135920731 | 3.62 |
ENSMUST00000030434.4
|
Fuca1
|
fucosidase, alpha-L- 1, tissue |
| chr17_+_43801823 | 3.61 |
ENSMUST00000044895.5
|
Rcan2
|
regulator of calcineurin 2 |
| chr11_+_102435285 | 3.60 |
ENSMUST00000125819.2
ENSMUST00000177428.1 |
Grn
|
granulin |
| chr11_+_99041237 | 3.59 |
ENSMUST00000017637.6
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr11_+_104231573 | 3.59 |
ENSMUST00000132977.1
ENSMUST00000132245.1 ENSMUST00000100347.4 |
Mapt
|
microtubule-associated protein tau |
| chr19_+_5038826 | 3.58 |
ENSMUST00000053705.6
|
B3gnt1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr4_-_124850670 | 3.57 |
ENSMUST00000163946.1
ENSMUST00000106190.3 |
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr10_+_7589788 | 3.57 |
ENSMUST00000134346.1
ENSMUST00000019931.5 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr13_+_64161862 | 3.49 |
ENSMUST00000021929.8
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr7_-_19149189 | 3.43 |
ENSMUST00000032566.1
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr17_+_35262730 | 3.41 |
ENSMUST00000172785.1
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr19_+_36554661 | 3.37 |
ENSMUST00000169036.2
ENSMUST00000047247.5 |
Hectd2
|
HECT domain containing 2 |
| chr17_+_32506446 | 3.36 |
ENSMUST00000165999.1
|
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr4_-_138367966 | 3.35 |
ENSMUST00000030535.3
|
Cda
|
cytidine deaminase |
| chr2_-_71750083 | 3.33 |
ENSMUST00000180494.1
|
Gm17250
|
predicted gene, 17250 |
| chr2_+_173153048 | 3.31 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr5_-_34187670 | 3.27 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr4_-_124850473 | 3.26 |
ENSMUST00000137769.2
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr4_+_152008803 | 3.25 |
ENSMUST00000097773.3
|
Klhl21
|
kelch-like 21 |
| chr18_+_12333953 | 3.22 |
ENSMUST00000092070.6
|
Lama3
|
laminin, alpha 3 |
| chr1_+_88200601 | 3.21 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr2_-_181039286 | 3.20 |
ENSMUST00000067120.7
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr9_-_97018823 | 3.19 |
ENSMUST00000055433.4
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr1_-_106759727 | 3.17 |
ENSMUST00000010049.4
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr6_-_147087023 | 3.12 |
ENSMUST00000100780.2
|
Mansc4
|
MANSC domain containing 4 |
| chr7_+_35119285 | 3.11 |
ENSMUST00000042985.9
|
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr2_+_18672384 | 3.09 |
ENSMUST00000171845.1
ENSMUST00000061158.4 |
Commd3
|
COMM domain containing 3 |
| chr5_-_140830430 | 3.08 |
ENSMUST00000000153.4
|
Gna12
|
guanine nucleotide binding protein, alpha 12 |
| chr7_+_16309577 | 3.08 |
ENSMUST00000002152.6
|
Bbc3
|
BCL2 binding component 3 |
| chr11_-_106487833 | 3.04 |
ENSMUST00000106801.1
|
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr2_-_181039160 | 3.02 |
ENSMUST00000108851.1
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr4_+_105157339 | 3.02 |
ENSMUST00000064139.7
|
Ppap2b
|
phosphatidic acid phosphatase type 2B |
| chr5_+_87000838 | 3.01 |
ENSMUST00000031186.7
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr2_+_25428699 | 3.01 |
ENSMUST00000102919.3
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr16_+_37868383 | 3.00 |
ENSMUST00000078717.6
|
Lrrc58
|
leucine rich repeat containing 58 |
| chr17_+_35470083 | 2.99 |
ENSMUST00000174525.1
ENSMUST00000068291.6 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr11_-_106487796 | 2.98 |
ENSMUST00000001059.2
ENSMUST00000106799.1 ENSMUST00000106800.1 |
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr10_+_75893398 | 2.95 |
ENSMUST00000009236.4
|
Derl3
|
Der1-like domain family, member 3 |
| chr2_+_23069210 | 2.94 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr4_+_144893077 | 2.94 |
ENSMUST00000154208.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr17_-_45595842 | 2.92 |
ENSMUST00000164618.1
ENSMUST00000097317.3 ENSMUST00000170113.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr2_-_164443177 | 2.91 |
ENSMUST00000017153.3
|
Sdc4
|
syndecan 4 |
| chr10_+_61695503 | 2.87 |
ENSMUST00000020284.4
|
Tysnd1
|
trypsin domain containing 1 |
| chr11_-_120784183 | 2.86 |
ENSMUST00000026156.7
|
Rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr10_-_127888688 | 2.85 |
ENSMUST00000047199.4
|
Rdh7
|
retinol dehydrogenase 7 |
| chr1_+_88095054 | 2.85 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr9_+_65265173 | 2.85 |
ENSMUST00000048762.1
|
Cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr3_+_95318782 | 2.85 |
ENSMUST00000139866.1
|
Cers2
|
ceramide synthase 2 |
| chr4_-_124850652 | 2.84 |
ENSMUST00000125776.1
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr5_-_92505518 | 2.83 |
ENSMUST00000031377.7
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr8_-_70857008 | 2.81 |
ENSMUST00000110081.3
ENSMUST00000110078.3 |
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr13_-_43480973 | 2.80 |
ENSMUST00000144326.2
|
Ranbp9
|
RAN binding protein 9 |
| chr19_-_6067785 | 2.80 |
ENSMUST00000162575.1
ENSMUST00000159084.1 ENSMUST00000161718.1 ENSMUST00000162810.1 ENSMUST00000025713.5 ENSMUST00000113543.2 ENSMUST00000161528.1 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr2_+_23069057 | 2.80 |
ENSMUST00000114526.1
ENSMUST00000114529.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr5_+_35041539 | 2.79 |
ENSMUST00000030985.6
|
Hgfac
|
hepatocyte growth factor activator |
| chr6_-_47813512 | 2.79 |
ENSMUST00000077290.7
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr17_+_26113286 | 2.77 |
ENSMUST00000025010.7
|
Tmem8
|
transmembrane protein 8 (five membrane-spanning domains) |
| chr2_-_91195097 | 2.77 |
ENSMUST00000002177.2
ENSMUST00000111354.1 |
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr17_-_32947372 | 2.75 |
ENSMUST00000139353.1
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chrX_+_100767719 | 2.74 |
ENSMUST00000000901.6
ENSMUST00000113736.2 ENSMUST00000087984.4 |
Dlg3
|
discs, large homolog 3 (Drosophila) |
| chr4_+_144892813 | 2.73 |
ENSMUST00000105744.1
ENSMUST00000171001.1 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr7_+_130936172 | 2.73 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr6_-_72617000 | 2.71 |
ENSMUST00000070524.4
|
Tgoln1
|
trans-golgi network protein |
| chr11_+_50602072 | 2.70 |
ENSMUST00000040523.8
|
Adamts2
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 |
| chr10_-_127351753 | 2.68 |
ENSMUST00000059718.4
|
Inhbe
|
inhibin beta E |
| chr7_+_101394361 | 2.67 |
ENSMUST00000154239.1
ENSMUST00000098243.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_32947389 | 2.67 |
ENSMUST00000075253.6
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr3_+_135826075 | 2.66 |
ENSMUST00000029810.5
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr6_-_11907419 | 2.65 |
ENSMUST00000031637.5
|
Ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr4_+_144893127 | 2.65 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr11_-_95587691 | 2.63 |
ENSMUST00000000122.6
|
Ngfr
|
nerve growth factor receptor (TNFR superfamily, member 16) |
| chr11_+_104231465 | 2.63 |
ENSMUST00000145227.1
|
Mapt
|
microtubule-associated protein tau |
| chr11_+_104231515 | 2.62 |
ENSMUST00000106993.3
|
Mapt
|
microtubule-associated protein tau |
| chr7_+_3694512 | 2.61 |
ENSMUST00000108627.3
|
Tsen34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae) |
| chr1_-_72212249 | 2.61 |
ENSMUST00000048860.7
|
Mreg
|
melanoregulin |
| chr17_-_36129425 | 2.61 |
ENSMUST00000046131.9
ENSMUST00000173322.1 ENSMUST00000172968.1 |
Gm7030
|
predicted gene 7030 |
| chr2_+_55437100 | 2.57 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr10_-_24101951 | 2.56 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr8_-_69890967 | 2.56 |
ENSMUST00000152938.1
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr9_+_6168638 | 2.55 |
ENSMUST00000058692.7
|
Pdgfd
|
platelet-derived growth factor, D polypeptide |
| chr11_-_115187827 | 2.55 |
ENSMUST00000103041.1
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr3_+_54735536 | 2.53 |
ENSMUST00000044567.3
|
Alg5
|
asparagine-linked glycosylation 5 (dolichyl-phosphate beta-glucosyltransferase) |
| chrX_-_112698642 | 2.53 |
ENSMUST00000039887.3
|
Pof1b
|
premature ovarian failure 1B |
| chr10_-_24836165 | 2.52 |
ENSMUST00000020169.7
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr19_+_4855129 | 2.52 |
ENSMUST00000119694.1
|
Ctsf
|
cathepsin F |
| chr17_+_35342242 | 2.51 |
ENSMUST00000074806.5
|
H2-Q2
|
histocompatibility 2, Q region locus 2 |
| chr16_+_93607831 | 2.51 |
ENSMUST00000039659.8
|
Cbr1
|
carbonyl reductase 1 |
| chr6_-_124863877 | 2.51 |
ENSMUST00000046893.7
|
Gpr162
|
G protein-coupled receptor 162 |
| chr1_-_82291370 | 2.49 |
ENSMUST00000069799.2
|
Irs1
|
insulin receptor substrate 1 |
| chr7_+_107370728 | 2.49 |
ENSMUST00000137663.1
ENSMUST00000073459.5 |
Syt9
|
synaptotagmin IX |
| chr8_-_84773381 | 2.49 |
ENSMUST00000109764.1
|
Nfix
|
nuclear factor I/X |
| chr16_-_10543028 | 2.49 |
ENSMUST00000184863.1
ENSMUST00000038281.5 |
Dexi
|
dexamethasone-induced transcript |
| chr15_-_74728011 | 2.49 |
ENSMUST00000023261.2
|
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr4_+_149545102 | 2.48 |
ENSMUST00000105692.1
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr17_+_87282880 | 2.48 |
ENSMUST00000041110.5
ENSMUST00000125875.1 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr9_-_54501496 | 2.47 |
ENSMUST00000118600.1
ENSMUST00000118163.1 |
Dmxl2
|
Dmx-like 2 |
| chr10_-_127621107 | 2.47 |
ENSMUST00000049149.8
|
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr11_-_99851608 | 2.47 |
ENSMUST00000107437.1
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr6_-_119330668 | 2.47 |
ENSMUST00000112756.1
|
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr7_-_27337667 | 2.46 |
ENSMUST00000038618.6
ENSMUST00000108369.2 |
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr5_+_102845007 | 2.45 |
ENSMUST00000070000.4
|
Arhgap24
|
Rho GTPase activating protein 24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 3.9 | 42.4 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 3.3 | 19.5 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 3.2 | 9.7 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 2.5 | 12.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 2.5 | 7.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.3 | 11.7 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 2.1 | 6.4 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 1.9 | 11.7 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.9 | 5.7 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 1.7 | 5.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.7 | 30.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 1.6 | 4.8 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.6 | 4.7 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.6 | 14.0 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 1.5 | 6.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.5 | 7.4 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.4 | 7.0 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.3 | 5.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 1.3 | 3.9 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.3 | 6.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 1.3 | 10.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 1.2 | 9.6 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.1 | 3.3 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.1 | 3.3 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.0 | 4.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 1.0 | 4.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.0 | 1.9 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.0 | 2.9 | GO:0009804 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.9 | 2.8 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.9 | 3.7 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.9 | 4.6 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.9 | 4.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.9 | 1.8 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.9 | 2.6 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.8 | 10.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.8 | 2.5 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.8 | 5.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.8 | 8.2 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.8 | 22.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.8 | 2.4 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.8 | 3.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.8 | 1.5 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.8 | 2.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.8 | 2.3 | GO:1904173 | regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.8 | 5.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.8 | 2.3 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.7 | 3.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.7 | 4.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.7 | 4.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.7 | 2.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.7 | 2.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.7 | 2.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.7 | 3.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.7 | 14.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.6 | 4.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.6 | 2.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.6 | 3.8 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.6 | 1.2 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.6 | 5.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.6 | 3.6 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.6 | 2.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.6 | 1.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.6 | 2.9 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.6 | 0.6 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.5 | 6.5 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.5 | 1.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.5 | 8.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 2.5 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.5 | 2.4 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.5 | 6.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 2.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.5 | 3.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.4 | 2.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 0.4 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.4 | 2.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.4 | 2.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 2.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 4.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.4 | 2.9 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 16.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.4 | 1.6 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.4 | 1.9 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.4 | 0.8 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.4 | 3.8 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.4 | 2.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.4 | 1.8 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.4 | 2.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 1.0 | GO:0060066 | oviduct development(GO:0060066) response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.3 | 13.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 6.8 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.3 | 1.7 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 1.7 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.3 | 1.0 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 1.0 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.3 | 1.3 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.3 | 3.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.3 | 5.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 3.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.3 | 1.9 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.3 | 2.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 | 3.6 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 1.5 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 | 3.2 | GO:0052697 | flavonoid metabolic process(GO:0009812) flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 2.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.3 | 0.9 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.3 | 5.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.3 | 1.4 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 4.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 3.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.3 | 2.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 1.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 2.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 5.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.3 | 0.8 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.3 | 2.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 9.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 0.8 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.3 | 2.7 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 1.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 0.8 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 0.8 | GO:0060220 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) negative regulation of immature T cell proliferation in thymus(GO:0033088) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.3 | 0.8 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.2 | 1.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 3.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.7 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 2.4 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 1.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 2.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 0.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 1.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 1.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.2 | 3.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 1.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 2.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 2.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 3.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 1.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 3.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 1.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 0.8 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 1.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.2 | 0.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 1.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 1.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 1.9 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.2 | 0.2 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.2 | 0.8 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.2 | 1.9 | GO:0098885 | maternal process involved in parturition(GO:0060137) modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.2 | 0.4 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.2 | 1.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 2.5 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.2 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.5 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 2.5 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.2 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 1.8 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 2.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.0 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 1.6 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.2 | 2.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 0.5 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.5 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 0.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.8 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 1.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.4 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 3.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.6 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 2.7 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 1.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.8 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.6 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 1.9 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 2.6 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 0.8 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 1.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 4.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.8 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.0 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.5 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 | 1.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.1 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 1.0 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 4.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 2.9 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 2.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.3 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.7 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 0.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.6 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 1.1 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.9 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 0.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 2.0 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 4.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.7 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 | 0.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 5.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.0 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 2.1 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 2.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 6.8 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 0.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 3.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.9 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 1.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.0 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 1.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.1 | GO:2000646 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 1.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 2.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.9 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 2.8 | GO:0009268 | response to pH(GO:0009268) |
| 0.1 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 1.5 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 1.2 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 2.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 3.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.3 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 1.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 2.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 2.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.9 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.5 | GO:0030953 | positive regulation of centrosome duplication(GO:0010825) astral microtubule organization(GO:0030953) |
| 0.0 | 1.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 1.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 6.3 | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032434) |
| 0.0 | 0.7 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 5.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 4.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 1.9 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.9 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 0.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 1.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 3.3 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.2 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.4 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 3.2 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.0 | GO:0045991 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 1.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.8 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 2.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.0 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.8 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.9 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 25.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.7 | 8.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 2.2 | 19.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 1.9 | 24.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.9 | 22.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.6 | 6.4 | GO:0060187 | cell pole(GO:0060187) |
| 1.1 | 3.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.0 | 3.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.9 | 6.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.8 | 2.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.8 | 2.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.7 | 9.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 1.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.6 | 12.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.6 | 4.9 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.6 | 7.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 2.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.5 | 1.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.5 | 9.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.5 | 1.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.5 | 3.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.4 | 9.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 1.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.4 | 6.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 2.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 1.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 1.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 1.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.3 | 3.3 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.3 | 7.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 1.7 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 11.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 0.8 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 2.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 1.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 3.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 4.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 1.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 2.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 1.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 2.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 1.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 1.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 1.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 2.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.6 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 6.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 4.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 3.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 2.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 2.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.5 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.1 | 2.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 6.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 3.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.5 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 1.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 5.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 3.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.4 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 8.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 8.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 4.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0061700 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.0 | 10.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 15.5 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 7.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 1.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 2.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.3 | GO:0005009 | insulin-activated receptor activity(GO:0005009) pheromone activity(GO:0005186) |
| 2.7 | 8.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 2.0 | 6.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 2.0 | 14.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.9 | 24.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.8 | 17.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.6 | 4.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.3 | 10.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.3 | 6.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 1.2 | 3.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.2 | 3.6 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.2 | 8.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.2 | 5.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.0 | 5.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.0 | 3.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.0 | 7.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.0 | 23.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.8 | 5.7 | GO:0002054 | nucleobase binding(GO:0002054) hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.8 | 3.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.8 | 18.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.7 | 13.5 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.7 | 9.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 10.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.7 | 2.8 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.7 | 4.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.6 | 4.5 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.6 | 12.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.6 | 2.5 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.6 | 22.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 3.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.6 | 3.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.6 | 1.8 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.6 | 1.7 | GO:0030350 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.6 | 3.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 4.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.5 | 2.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.5 | 1.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.5 | 2.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.5 | 1.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.5 | 2.3 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.4 | 2.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.4 | 2.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 2.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 2.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 1.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.4 | 1.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 2.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.4 | 1.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.4 | 2.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.4 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 6.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 1.8 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.4 | 3.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.4 | 3.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 1.0 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 2.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 1.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 12.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 1.0 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.3 | 1.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 1.2 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 14.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 2.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 1.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.3 | 2.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 1.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 5.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 4.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.3 | 3.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.3 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 4.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 7.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 0.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 14.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 1.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 0.9 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.2 | 4.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 2.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 2.8 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.2 | 1.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 1.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 2.7 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 0.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 3.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 4.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 5.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 1.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 7.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 3.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 3.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 2.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 3.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 0.8 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 3.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 5.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 0.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 2.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 3.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 3.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 5.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.5 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.8 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 3.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 2.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.8 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 3.0 | GO:1905030 | voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 2.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.9 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 1.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 8.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 3.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 2.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 2.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 2.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0051431 | histone deacetylase regulator activity(GO:0035033) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 5.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.5 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.7 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 3.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 16.4 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.1 | 1.0 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 5.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 2.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 18.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 3.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.3 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 1.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.6 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 2.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 2.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 6.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 4.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 9.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 3.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 22.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 21.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.4 | 16.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 2.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 5.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 7.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 10.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 6.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 10.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 9.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 5.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 3.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 6.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 1.0 | 29.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.0 | 12.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.0 | 13.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.7 | 6.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 5.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.6 | 11.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 9.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 19.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 5.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 8.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 4.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 8.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.4 | 34.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 3.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 4.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 5.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 4.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.3 | 4.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 3.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 3.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 7.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 3.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 1.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 8.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.2 | 1.2 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.2 | 3.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 1.3 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.2 | 3.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 5.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 4.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 7.1 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.1 | 1.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 6.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 5.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 9.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.2 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 2.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 8.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.6 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |