Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
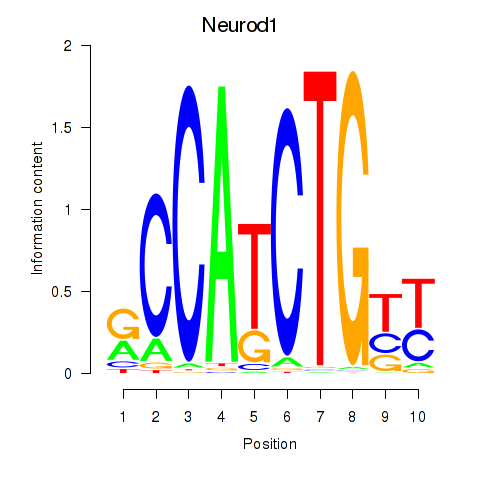
Results for Neurod1
Z-value: 1.15
Transcription factors associated with Neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod1
|
ENSMUSG00000034701.9 | neurogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurod1 | mm10_v2_chr2_-_79456750_79456761 | -0.33 | 4.8e-02 | Click! |
Activity profile of Neurod1 motif
Sorted Z-values of Neurod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127866457 | 10.54 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr10_-_109010955 | 8.07 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr10_+_127801145 | 7.94 |
ENSMUST00000071646.1
|
Rdh16
|
retinol dehydrogenase 16 |
| chr2_-_28563362 | 7.80 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr4_-_137409777 | 7.74 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr4_-_137430517 | 6.92 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr11_+_48837465 | 6.41 |
ENSMUST00000046903.5
|
Trim7
|
tripartite motif-containing 7 |
| chr19_+_52264323 | 6.21 |
ENSMUST00000039652.4
|
Ins1
|
insulin I |
| chr7_-_142679533 | 6.17 |
ENSMUST00000162317.1
ENSMUST00000125933.1 ENSMUST00000105931.1 ENSMUST00000105930.1 ENSMUST00000105933.1 ENSMUST00000105932.1 ENSMUST00000000220.2 |
Ins2
|
insulin II |
| chr18_+_45268876 | 5.32 |
ENSMUST00000183850.1
ENSMUST00000066890.7 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr10_+_127759780 | 5.04 |
ENSMUST00000128247.1
|
RP23-386P10.11
|
Protein Rdh9 |
| chr11_-_5950018 | 4.75 |
ENSMUST00000102920.3
|
Gck
|
glucokinase |
| chr1_+_167598450 | 4.27 |
ENSMUST00000111386.1
ENSMUST00000111384.1 |
Rxrg
|
retinoid X receptor gamma |
| chr1_+_167598384 | 3.91 |
ENSMUST00000015987.3
|
Rxrg
|
retinoid X receptor gamma |
| chr7_+_27119909 | 3.80 |
ENSMUST00000003100.8
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr1_+_72824482 | 3.73 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr17_+_36942910 | 3.63 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr11_+_117809653 | 3.61 |
ENSMUST00000026649.7
ENSMUST00000177131.1 ENSMUST00000132298.1 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr19_-_42202150 | 3.55 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr3_+_14863495 | 3.43 |
ENSMUST00000029076.4
|
Car3
|
carbonic anhydrase 3 |
| chr6_+_124570294 | 3.10 |
ENSMUST00000184647.1
|
C1rb
|
complement component 1, r subcomponent B |
| chr16_+_42907563 | 3.04 |
ENSMUST00000151244.1
ENSMUST00000114694.2 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_-_114562945 | 2.99 |
ENSMUST00000119712.1
ENSMUST00000032908.8 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr5_-_151369172 | 2.97 |
ENSMUST00000067770.3
|
D730045B01Rik
|
RIKEN cDNA D730045B01 gene |
| chr10_+_127776374 | 2.95 |
ENSMUST00000136223.1
ENSMUST00000052652.6 |
Rdh9
|
retinol dehydrogenase 9 |
| chr17_+_36943025 | 2.88 |
ENSMUST00000173072.1
|
Rnf39
|
ring finger protein 39 |
| chr8_+_45507768 | 2.82 |
ENSMUST00000067065.7
ENSMUST00000098788.3 ENSMUST00000067107.7 ENSMUST00000171337.2 ENSMUST00000138049.1 ENSMUST00000141039.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr14_-_57104693 | 2.67 |
ENSMUST00000055698.7
|
Gjb2
|
gap junction protein, beta 2 |
| chr18_-_74961252 | 2.66 |
ENSMUST00000066532.4
|
Lipg
|
lipase, endothelial |
| chr10_-_127888688 | 2.60 |
ENSMUST00000047199.4
|
Rdh7
|
retinol dehydrogenase 7 |
| chr16_+_56204313 | 2.59 |
ENSMUST00000160116.1
ENSMUST00000069936.7 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr1_-_173367638 | 2.54 |
ENSMUST00000005470.4
ENSMUST00000111220.1 |
Cadm3
|
cell adhesion molecule 3 |
| chr4_-_141623799 | 2.52 |
ENSMUST00000038661.7
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr7_+_28071230 | 2.50 |
ENSMUST00000138392.1
ENSMUST00000076648.7 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chrX_+_139800795 | 2.47 |
ENSMUST00000054889.3
|
Cldn2
|
claudin 2 |
| chr14_+_123659971 | 2.47 |
ENSMUST00000049681.7
|
Itgbl1
|
integrin, beta-like 1 |
| chr9_+_44066993 | 2.39 |
ENSMUST00000034508.7
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr2_-_52558539 | 2.30 |
ENSMUST00000102760.3
ENSMUST00000102761.2 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr17_-_56005566 | 2.27 |
ENSMUST00000043785.6
|
Stap2
|
signal transducing adaptor family member 2 |
| chr11_+_115462464 | 2.26 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr7_-_19796789 | 2.16 |
ENSMUST00000108449.2
ENSMUST00000043822.7 |
Cblc
|
Casitas B-lineage lymphoma c |
| chr18_-_61911783 | 2.09 |
ENSMUST00000049378.8
ENSMUST00000166783.1 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr10_+_107271827 | 2.08 |
ENSMUST00000020057.8
ENSMUST00000105280.3 |
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr11_-_35980473 | 2.05 |
ENSMUST00000018993.6
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr16_-_46010212 | 2.05 |
ENSMUST00000130481.1
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr17_-_12851893 | 2.05 |
ENSMUST00000162389.1
ENSMUST00000162119.1 ENSMUST00000159223.1 |
Mas1
|
MAS1 oncogene |
| chr8_+_119394866 | 2.02 |
ENSMUST00000098367.4
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr7_-_141437829 | 2.01 |
ENSMUST00000019226.7
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr5_+_151368683 | 1.99 |
ENSMUST00000181114.1
ENSMUST00000181555.1 |
1700028E10Rik
|
RIKEN cDNA 1700028E10 gene |
| chr2_-_160872985 | 1.98 |
ENSMUST00000109460.1
ENSMUST00000127201.1 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr17_+_46254017 | 1.97 |
ENSMUST00000095262.4
|
Lrrc73
|
leucine rich repeat containing 73 |
| chr6_-_87690819 | 1.97 |
ENSMUST00000162547.1
|
1810020O05Rik
|
Riken cDNA 1810020O05 gene |
| chr7_-_141437587 | 1.93 |
ENSMUST00000172654.1
ENSMUST00000106006.1 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr19_-_24861828 | 1.89 |
ENSMUST00000047666.4
|
Pgm5
|
phosphoglucomutase 5 |
| chr9_+_44067072 | 1.88 |
ENSMUST00000177054.1
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr1_+_131970589 | 1.85 |
ENSMUST00000027695.6
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr11_-_53773187 | 1.85 |
ENSMUST00000170390.1
|
Gm17334
|
predicted gene, 17334 |
| chr10_+_127759721 | 1.84 |
ENSMUST00000073639.5
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr2_-_84743655 | 1.80 |
ENSMUST00000181711.1
|
Gm19426
|
predicted gene, 19426 |
| chr10_+_127849917 | 1.75 |
ENSMUST00000077530.2
|
Rdh19
|
retinol dehydrogenase 19 |
| chr7_+_101394361 | 1.74 |
ENSMUST00000154239.1
ENSMUST00000098243.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_+_56005672 | 1.71 |
ENSMUST00000133998.1
|
Mpnd
|
MPN domain containing |
| chr10_+_60106198 | 1.69 |
ENSMUST00000121820.2
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr2_-_29253001 | 1.61 |
ENSMUST00000071201.4
|
Ntng2
|
netrin G2 |
| chr9_-_50746501 | 1.60 |
ENSMUST00000034564.1
|
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr7_+_100009914 | 1.58 |
ENSMUST00000107084.1
|
Chrdl2
|
chordin-like 2 |
| chr9_+_100643605 | 1.57 |
ENSMUST00000041418.6
|
Stag1
|
stromal antigen 1 |
| chr5_-_86197846 | 1.57 |
ENSMUST00000094654.2
|
Gnrhr
|
gonadotropin releasing hormone receptor |
| chr18_+_84088077 | 1.56 |
ENSMUST00000060223.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr2_-_84775420 | 1.55 |
ENSMUST00000111641.1
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr2_-_84775388 | 1.55 |
ENSMUST00000023994.3
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr15_-_76090013 | 1.54 |
ENSMUST00000019516.4
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr9_+_100643755 | 1.52 |
ENSMUST00000133388.1
|
Stag1
|
stromal antigen 1 |
| chr3_-_84259812 | 1.51 |
ENSMUST00000107691.1
|
Trim2
|
tripartite motif-containing 2 |
| chr2_-_52335134 | 1.51 |
ENSMUST00000075301.3
|
Neb
|
nebulin |
| chr10_-_95415484 | 1.51 |
ENSMUST00000172070.1
ENSMUST00000150432.1 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr2_+_96318014 | 1.51 |
ENSMUST00000135431.1
ENSMUST00000162807.2 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr9_-_63602417 | 1.51 |
ENSMUST00000171243.1
ENSMUST00000163982.1 ENSMUST00000163624.1 |
Iqch
|
IQ motif containing H |
| chr7_+_27607997 | 1.50 |
ENSMUST00000142365.1
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr8_-_119635553 | 1.48 |
ENSMUST00000061828.3
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr7_-_90129339 | 1.47 |
ENSMUST00000181189.1
|
2310010J17Rik
|
RIKEN cDNA 2310010J17 gene |
| chr5_+_90460889 | 1.46 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr7_-_98361275 | 1.45 |
ENSMUST00000094161.4
ENSMUST00000164726.1 ENSMUST00000167405.1 |
Tsku
|
tsukushi |
| chr6_-_136171722 | 1.44 |
ENSMUST00000053880.6
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr8_+_127064107 | 1.43 |
ENSMUST00000162536.1
ENSMUST00000026921.6 ENSMUST00000162665.1 ENSMUST00000160766.1 ENSMUST00000162602.1 ENSMUST00000162531.1 ENSMUST00000160581.1 ENSMUST00000161355.1 ENSMUST00000159537.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr9_-_63602464 | 1.43 |
ENSMUST00000080527.5
ENSMUST00000042322.4 |
Iqch
|
IQ motif containing H |
| chr19_+_4082473 | 1.42 |
ENSMUST00000159148.1
|
Cabp2
|
calcium binding protein 2 |
| chr9_-_105960642 | 1.42 |
ENSMUST00000165165.2
|
Col6a5
|
collagen, type VI, alpha 5 |
| chr15_+_25622525 | 1.40 |
ENSMUST00000110457.1
ENSMUST00000137601.1 |
Myo10
|
myosin X |
| chr19_+_57611020 | 1.37 |
ENSMUST00000077282.5
|
Atrnl1
|
attractin like 1 |
| chr18_-_77565050 | 1.35 |
ENSMUST00000182153.1
ENSMUST00000182146.1 ENSMUST00000026494.7 ENSMUST00000182024.1 |
Rnf165
|
ring finger protein 165 |
| chr4_+_80910646 | 1.35 |
ENSMUST00000055922.3
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr6_-_97252779 | 1.34 |
ENSMUST00000095655.2
|
Lmod3
|
leiomodin 3 (fetal) |
| chr1_-_91413163 | 1.27 |
ENSMUST00000086851.1
|
Hes6
|
hairy and enhancer of split 6 |
| chr2_-_10130638 | 1.27 |
ENSMUST00000042290.7
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr11_+_97030130 | 1.27 |
ENSMUST00000153482.1
|
Scrn2
|
secernin 2 |
| chr6_+_56017489 | 1.27 |
ENSMUST00000052827.4
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr7_-_98361310 | 1.26 |
ENSMUST00000165257.1
|
Tsku
|
tsukushi |
| chr17_+_34670535 | 1.24 |
ENSMUST00000168533.1
ENSMUST00000087399.4 |
Tnxb
|
tenascin XB |
| chr1_-_192834719 | 1.24 |
ENSMUST00000057543.2
|
A730013G03Rik
|
RIKEN cDNA A730013G03 gene |
| chr3_+_96246685 | 1.23 |
ENSMUST00000176059.1
ENSMUST00000177796.1 |
Hist2h3c1
|
histone cluster 2, H3c1 |
| chr1_+_162639148 | 1.22 |
ENSMUST00000028020.9
|
Myoc
|
myocilin |
| chr13_-_60177357 | 1.18 |
ENSMUST00000065086.4
|
Gas1
|
growth arrest specific 1 |
| chr11_+_97029925 | 1.18 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr1_+_162570515 | 1.15 |
ENSMUST00000132158.1
ENSMUST00000135241.1 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr7_-_45061706 | 1.15 |
ENSMUST00000107832.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr9_+_100643448 | 1.14 |
ENSMUST00000146312.1
ENSMUST00000129269.1 |
Stag1
|
stromal antigen 1 |
| chr6_+_54264839 | 1.13 |
ENSMUST00000146114.1
|
Chn2
|
chimerin (chimaerin) 2 |
| chr18_-_12819842 | 1.13 |
ENSMUST00000119043.1
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr11_+_117809687 | 1.12 |
ENSMUST00000120928.1
ENSMUST00000175737.1 |
Syngr2
|
synaptogyrin 2 |
| chr7_+_128523576 | 1.12 |
ENSMUST00000033136.7
|
Bag3
|
BCL2-associated athanogene 3 |
| chr19_+_5038826 | 1.10 |
ENSMUST00000053705.6
|
B3gnt1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr7_+_27607748 | 1.10 |
ENSMUST00000136962.1
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr10_-_128922888 | 1.09 |
ENSMUST00000135161.1
|
Rdh5
|
retinol dehydrogenase 5 |
| chr9_-_107710475 | 1.07 |
ENSMUST00000080560.3
|
Sema3f
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr11_+_67798269 | 1.07 |
ENSMUST00000168612.1
ENSMUST00000040574.4 |
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr10_+_79973737 | 1.06 |
ENSMUST00000149148.1
|
Grin3b
|
glutamate receptor, ionotropic, NMDA3B |
| chr7_-_45870928 | 1.05 |
ENSMUST00000146672.1
|
Grin2d
|
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
| chr19_-_32712287 | 1.05 |
ENSMUST00000070210.4
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chrX_-_48208566 | 1.04 |
ENSMUST00000037960.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr7_+_140704165 | 1.04 |
ENSMUST00000080681.3
|
Olfr541
|
olfactory receptor 541 |
| chr16_+_84774123 | 1.03 |
ENSMUST00000114195.1
|
Jam2
|
junction adhesion molecule 2 |
| chr11_+_78503449 | 1.03 |
ENSMUST00000001130.6
ENSMUST00000125670.2 |
Sebox
|
SEBOX homeobox |
| chr11_+_70030023 | 1.02 |
ENSMUST00000143920.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr11_-_116131073 | 1.01 |
ENSMUST00000106440.2
ENSMUST00000067632.3 |
Trim65
|
tripartite motif-containing 65 |
| chr4_-_22488296 | 1.00 |
ENSMUST00000178174.1
|
Pou3f2
|
POU domain, class 3, transcription factor 2 |
| chr5_-_143138200 | 1.00 |
ENSMUST00000164536.2
|
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chr11_-_99556846 | 0.99 |
ENSMUST00000092699.2
|
Krtap3-2
|
keratin associated protein 3-2 |
| chr11_-_119086221 | 0.98 |
ENSMUST00000026665.7
|
Cbx4
|
chromobox 4 |
| chr4_-_25281801 | 0.97 |
ENSMUST00000102994.3
|
Ufl1
|
UFM1 specific ligase 1 |
| chr3_-_53863764 | 0.97 |
ENSMUST00000122330.1
ENSMUST00000146598.1 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr4_-_129227883 | 0.97 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr7_-_45061651 | 0.96 |
ENSMUST00000007981.3
ENSMUST00000107831.1 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr10_-_92375367 | 0.96 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr2_+_69219971 | 0.95 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr15_+_8968389 | 0.94 |
ENSMUST00000053308.9
ENSMUST00000166524.1 |
Ranbp3l
|
RAN binding protein 3-like |
| chr5_-_142608785 | 0.93 |
ENSMUST00000037048.7
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chrX_-_48208870 | 0.91 |
ENSMUST00000088935.3
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr11_+_109485606 | 0.91 |
ENSMUST00000106697.1
|
Arsg
|
arylsulfatase G |
| chr5_-_28210168 | 0.91 |
ENSMUST00000117098.1
|
Cnpy1
|
canopy 1 homolog (zebrafish) |
| chr9_+_78175898 | 0.90 |
ENSMUST00000180974.1
|
C920006O11Rik
|
RIKEN cDNA C920006O11 gene |
| chr13_+_51846673 | 0.90 |
ENSMUST00000021903.2
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr7_+_121707189 | 0.89 |
ENSMUST00000065310.2
|
1700069B07Rik
|
RIKEN cDNA 1700069B07 gene |
| chr1_-_180483410 | 0.88 |
ENSMUST00000136521.1
ENSMUST00000179826.1 |
6330403A02Rik
|
RIKEN cDNA 6330403A02 gene |
| chr3_-_73708399 | 0.88 |
ENSMUST00000029367.5
|
Bche
|
butyrylcholinesterase |
| chr13_-_22219820 | 0.87 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr11_-_98329641 | 0.86 |
ENSMUST00000041685.6
|
Neurod2
|
neurogenic differentiation 2 |
| chr2_+_163506808 | 0.85 |
ENSMUST00000143911.1
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr10_+_63100156 | 0.85 |
ENSMUST00000044059.3
|
Atoh7
|
atonal homolog 7 (Drosophila) |
| chr8_-_119635346 | 0.84 |
ENSMUST00000164382.1
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr5_-_74677792 | 0.84 |
ENSMUST00000117525.1
ENSMUST00000153543.1 ENSMUST00000039744.6 ENSMUST00000113531.2 ENSMUST00000121690.1 |
Lnx1
|
ligand of numb-protein X 1 |
| chr8_-_107065632 | 0.83 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr10_-_95415283 | 0.82 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr7_-_4970961 | 0.81 |
ENSMUST00000144863.1
|
Gm1078
|
predicted gene 1078 |
| chr7_-_142229971 | 0.81 |
ENSMUST00000097942.2
|
Krtap5-5
|
keratin associated protein 5-5 |
| chr7_-_140163014 | 0.80 |
ENSMUST00000050585.5
|
Olfr522
|
olfactory receptor 522 |
| chr4_-_148149684 | 0.80 |
ENSMUST00000126615.1
|
Fbxo6
|
F-box protein 6 |
| chr15_+_99099412 | 0.78 |
ENSMUST00000061295.6
|
Dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr4_-_20778852 | 0.77 |
ENSMUST00000102998.3
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr3_+_135438722 | 0.77 |
ENSMUST00000166033.1
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_118762607 | 0.77 |
ENSMUST00000059997.8
|
A430105I19Rik
|
RIKEN cDNA A430105I19 gene |
| chr7_-_28302238 | 0.76 |
ENSMUST00000108315.3
|
Dll3
|
delta-like 3 (Drosophila) |
| chr3_-_88254706 | 0.75 |
ENSMUST00000171887.1
|
Rhbg
|
Rhesus blood group-associated B glycoprotein |
| chr6_+_97807014 | 0.75 |
ENSMUST00000043637.7
|
Mitf
|
microphthalmia-associated transcription factor |
| chr15_+_101115738 | 0.75 |
ENSMUST00000070875.6
|
Ankrd33
|
ankyrin repeat domain 33 |
| chr7_-_140401044 | 0.74 |
ENSMUST00000080153.1
|
Olfr531
|
olfactory receptor 531 |
| chrX_-_20291776 | 0.74 |
ENSMUST00000072451.4
|
Slc9a7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
| chr11_+_67774608 | 0.74 |
ENSMUST00000181566.1
|
Gm12302
|
predicted gene 12302 |
| chr14_+_70530819 | 0.74 |
ENSMUST00000047331.6
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr4_-_42168603 | 0.74 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr10_+_80142295 | 0.73 |
ENSMUST00000003156.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr11_+_102836296 | 0.73 |
ENSMUST00000021302.8
ENSMUST00000107072.1 |
Higd1b
|
HIG1 domain family, member 1B |
| chr2_-_103372725 | 0.73 |
ENSMUST00000139065.1
|
A930006I01Rik
|
RIKEN cDNA A930006I01 gene |
| chr17_-_14203695 | 0.73 |
ENSMUST00000053218.5
|
Dact2
|
dapper homolog 2, antagonist of beta-catenin (xenopus) |
| chr11_+_72607221 | 0.73 |
ENSMUST00000021148.6
ENSMUST00000138247.1 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr10_-_121476248 | 0.73 |
ENSMUST00000026902.7
|
Rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr18_-_77186257 | 0.72 |
ENSMUST00000097520.2
|
Gm7276
|
predicted gene 7276 |
| chrX_-_36645359 | 0.72 |
ENSMUST00000051906.6
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr8_-_71381907 | 0.72 |
ENSMUST00000002466.8
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr1_+_87205799 | 0.71 |
ENSMUST00000027470.7
|
Chrng
|
cholinergic receptor, nicotinic, gamma polypeptide |
| chr1_+_143640664 | 0.71 |
ENSMUST00000038252.2
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_-_174438996 | 0.71 |
ENSMUST00000016400.8
|
Ctsz
|
cathepsin Z |
| chr7_-_102065044 | 0.70 |
ENSMUST00000130074.1
ENSMUST00000131104.1 ENSMUST00000096639.5 |
Rnf121
|
ring finger protein 121 |
| chr10_+_127420334 | 0.70 |
ENSMUST00000171434.1
|
R3hdm2
|
R3H domain containing 2 |
| chr2_-_46442681 | 0.70 |
ENSMUST00000123911.1
|
Gm13470
|
predicted gene 13470 |
| chr11_+_99879187 | 0.68 |
ENSMUST00000078442.3
|
Gm11567
|
predicted gene 11567 |
| chr5_+_30232581 | 0.68 |
ENSMUST00000145167.1
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr10_+_67538563 | 0.67 |
ENSMUST00000127820.1
|
Egr2
|
early growth response 2 |
| chr5_-_28210022 | 0.67 |
ENSMUST00000118882.1
|
Cnpy1
|
canopy 1 homolog (zebrafish) |
| chr17_+_8434423 | 0.67 |
ENSMUST00000074667.2
|
T
|
brachyury |
| chr7_+_3694512 | 0.67 |
ENSMUST00000108627.3
|
Tsen34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae) |
| chr4_-_25281752 | 0.67 |
ENSMUST00000038705.7
|
Ufl1
|
UFM1 specific ligase 1 |
| chr3_+_86084434 | 0.67 |
ENSMUST00000107664.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_-_173276144 | 0.67 |
ENSMUST00000139306.1
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_+_126034967 | 0.65 |
ENSMUST00000110442.1
|
Fgf7
|
fibroblast growth factor 7 |
| chr10_+_61175206 | 0.65 |
ENSMUST00000079235.5
|
Tbata
|
thymus, brain and testes associated |
| chr7_+_127471009 | 0.64 |
ENSMUST00000133938.1
|
Prr14
|
proline rich 14 |
| chr7_-_19861299 | 0.64 |
ENSMUST00000014830.7
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr3_+_9250602 | 0.64 |
ENSMUST00000155203.1
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr2_-_90022064 | 0.63 |
ENSMUST00000099758.1
|
Olfr1264
|
olfactory receptor 1264 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:1990535 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 1.6 | 10.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 3.5 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.0 | 3.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.0 | 3.8 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.9 | 4.7 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.8 | 5.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.7 | 8.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.7 | 2.0 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.7 | 2.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.6 | 7.8 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 2.7 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.5 | 1.9 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 2.6 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 2.6 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 2.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 1.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 1.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 1.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.3 | 2.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 0.9 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.3 | 0.9 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 0.8 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.3 | 1.1 | GO:0021664 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.3 | 0.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 2.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 1.0 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 | 0.9 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 2.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.7 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) |
| 0.2 | 8.2 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 0.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 1.7 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 1.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.6 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) negative regulation of sensory perception of pain(GO:1904057) |
| 0.2 | 4.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 0.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.7 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.2 | 0.7 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 | 1.1 | GO:0097490 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 2.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 0.7 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 1.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 4.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 2.7 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 2.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.4 | GO:0002194 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.6 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 1.6 | GO:0090003 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.1 | 4.1 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 2.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.5 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.0 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.6 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 0.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 2.3 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.2 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.7 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 1.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.3 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 1.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 3.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.4 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 1.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 3.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.6 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.1 | 0.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.7 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.8 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 2.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 6.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 3.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.5 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.8 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.7 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.7 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.7 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 6.1 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 2.2 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 2.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 3.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0043585 | retinol transport(GO:0034633) nose morphogenesis(GO:0043585) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.0 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 1.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.4 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.4 | 9.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 1.9 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.3 | 2.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 4.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 7.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 1.0 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 6.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 0.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 0.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 9.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 1.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 2.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 1.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 2.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 2.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 2.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 5.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 0.3 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 4.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 3.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 8.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 2.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 5.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 2.0 | 8.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.3 | 5.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 19.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.8 | 8.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.6 | 1.8 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.5 | 1.6 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.5 | 1.6 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.5 | 2.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 1.9 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 2.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 4.7 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.4 | 3.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 2.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 3.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 1.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 3.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 2.0 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.3 | 3.4 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.3 | 0.8 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.3 | 1.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 5.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 0.9 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.2 | 12.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 0.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 1.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 2.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.5 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.2 | 1.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 3.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 3.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 4.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.8 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 1.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 2.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 3.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 3.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 2.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 8.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 2.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 7.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 7.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 8.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 4.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 4.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 1.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 7.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 3.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 8.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 6.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |