Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
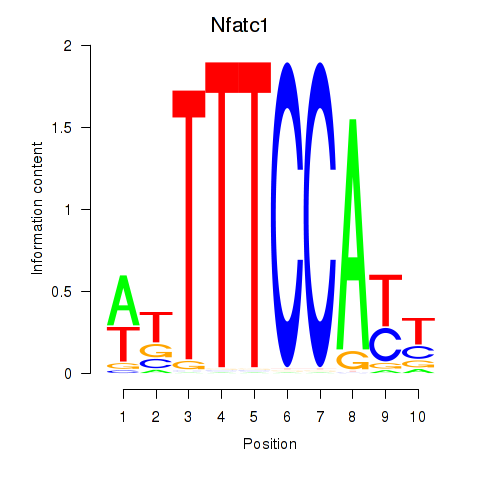
Results for Nfatc1
Z-value: 1.26
Transcription factors associated with Nfatc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc1
|
ENSMUSG00000033016.9 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm10_v2_chr18_-_80713062_80713080 | 0.37 | 2.6e-02 | Click! |
Activity profile of Nfatc1 motif
Sorted Z-values of Nfatc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_3023547 | 7.14 |
ENSMUST00000099046.3
|
Gm10718
|
predicted gene 10718 |
| chr9_+_3000922 | 6.79 |
ENSMUST00000151376.2
|
Gm10722
|
predicted gene 10722 |
| chr2_-_98667264 | 6.50 |
ENSMUST00000099683.1
|
Gm10800
|
predicted gene 10800 |
| chr9_+_3027439 | 6.21 |
ENSMUST00000177875.1
ENSMUST00000179982.1 |
Gm10717
|
predicted gene 10717 |
| chr16_-_16863817 | 6.02 |
ENSMUST00000124890.1
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr9_+_3013140 | 5.88 |
ENSMUST00000143083.2
|
Gm10721
|
predicted gene 10721 |
| chr7_-_142679533 | 5.39 |
ENSMUST00000162317.1
ENSMUST00000125933.1 ENSMUST00000105931.1 ENSMUST00000105930.1 ENSMUST00000105933.1 ENSMUST00000105932.1 ENSMUST00000000220.2 |
Ins2
|
insulin II |
| chr9_+_3017408 | 4.95 |
ENSMUST00000099049.3
|
Gm10719
|
predicted gene 10719 |
| chr9_+_3034599 | 4.74 |
ENSMUST00000178641.1
|
Gm17535
|
predicted gene, 17535 |
| chr19_+_52264323 | 4.65 |
ENSMUST00000039652.4
|
Ins1
|
insulin I |
| chr4_+_134864536 | 4.35 |
ENSMUST00000030627.7
|
Rhd
|
Rh blood group, D antigen |
| chr9_+_3025417 | 4.22 |
ENSMUST00000075573.6
|
Gm10717
|
predicted gene 10717 |
| chr14_-_19418930 | 4.09 |
ENSMUST00000177817.1
|
Gm21738
|
predicted gene, 21738 |
| chr9_+_3015654 | 4.09 |
ENSMUST00000099050.3
|
Gm10720
|
predicted gene 10720 |
| chr2_+_157560078 | 4.08 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr9_+_3005125 | 3.95 |
ENSMUST00000179881.1
|
Gm11168
|
predicted gene 11168 |
| chr9_+_3037111 | 3.88 |
ENSMUST00000177969.1
|
Gm10715
|
predicted gene 10715 |
| chr9_+_3004457 | 3.74 |
ENSMUST00000178348.1
|
Gm11168
|
predicted gene 11168 |
| chr7_-_134232005 | 3.25 |
ENSMUST00000134504.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr9_+_3036877 | 3.04 |
ENSMUST00000155807.2
|
Gm10715
|
predicted gene 10715 |
| chr4_-_44710408 | 3.04 |
ENSMUST00000134968.2
ENSMUST00000173821.1 ENSMUST00000174319.1 ENSMUST00000173733.1 ENSMUST00000172866.1 ENSMUST00000165417.2 ENSMUST00000107825.2 ENSMUST00000102932.3 ENSMUST00000107827.2 ENSMUST00000107826.2 |
Pax5
|
paired box gene 5 |
| chr9_+_3018753 | 3.02 |
ENSMUST00000179272.1
|
Gm10719
|
predicted gene 10719 |
| chr1_-_82768449 | 2.98 |
ENSMUST00000027331.2
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr16_+_45610380 | 2.96 |
ENSMUST00000161347.2
ENSMUST00000023339.4 |
Gcsam
|
germinal center associated, signaling and motility |
| chr14_+_118787894 | 2.95 |
ENSMUST00000047761.6
ENSMUST00000071546.7 |
Cldn10
|
claudin 10 |
| chr7_+_110773658 | 2.87 |
ENSMUST00000143786.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr7_+_110774240 | 2.78 |
ENSMUST00000147587.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr2_+_98662227 | 2.78 |
ENSMUST00000099684.3
|
Gm10801
|
predicted gene 10801 |
| chr7_+_30423266 | 2.70 |
ENSMUST00000046177.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr7_+_99535652 | 2.63 |
ENSMUST00000032995.8
ENSMUST00000162404.1 |
Arrb1
|
arrestin, beta 1 |
| chr15_-_79164477 | 2.61 |
ENSMUST00000040019.4
|
Sox10
|
SRY-box containing gene 10 |
| chrX_-_53240101 | 2.60 |
ENSMUST00000074861.2
|
Plac1
|
placental specific protein 1 |
| chr3_+_114030532 | 2.59 |
ENSMUST00000123619.1
ENSMUST00000092155.5 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr1_-_66935333 | 2.46 |
ENSMUST00000120415.1
ENSMUST00000119429.1 |
Myl1
|
myosin, light polypeptide 1 |
| chr6_+_30723541 | 2.45 |
ENSMUST00000115127.1
|
Mest
|
mesoderm specific transcript |
| chr6_+_41521782 | 2.45 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr4_-_88033328 | 2.40 |
ENSMUST00000078090.5
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chrX_+_139210031 | 2.37 |
ENSMUST00000113043.1
ENSMUST00000169886.1 ENSMUST00000113045.2 |
Mum1l1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr5_-_148392810 | 2.33 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr10_-_19851459 | 2.32 |
ENSMUST00000059805.4
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr2_-_170406501 | 2.25 |
ENSMUST00000154650.1
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr7_-_30821139 | 2.23 |
ENSMUST00000163504.1
|
Ffar2
|
free fatty acid receptor 2 |
| chr16_-_22161450 | 2.20 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr17_-_48432723 | 2.15 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr4_-_117125618 | 2.10 |
ENSMUST00000183310.1
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr12_-_40248073 | 2.07 |
ENSMUST00000169926.1
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr1_+_107535508 | 2.05 |
ENSMUST00000182198.1
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr6_-_41035501 | 2.01 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr15_-_66801577 | 2.00 |
ENSMUST00000168589.1
|
Sla
|
src-like adaptor |
| chr9_-_39604124 | 2.00 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr7_+_99535439 | 1.90 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chr2_+_35282380 | 1.89 |
ENSMUST00000028239.6
|
Gsn
|
gelsolin |
| chr6_-_142804390 | 1.87 |
ENSMUST00000111768.1
|
Gm766
|
predicted gene 766 |
| chr9_+_124102110 | 1.85 |
ENSMUST00000168841.1
ENSMUST00000055918.6 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr4_-_49593875 | 1.85 |
ENSMUST00000151542.1
|
Tmem246
|
transmembrane protein 246 |
| chr10_+_115817247 | 1.84 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr15_-_34356421 | 1.82 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chrX_-_133688978 | 1.69 |
ENSMUST00000149154.1
ENSMUST00000167944.1 |
Pcdh19
|
protocadherin 19 |
| chrX_-_9469288 | 1.64 |
ENSMUST00000015484.3
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr10_-_19014549 | 1.60 |
ENSMUST00000146388.1
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chrX_+_159627534 | 1.59 |
ENSMUST00000073094.3
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_+_146150174 | 1.56 |
ENSMUST00000098524.4
|
Mcoln2
|
mucolipin 2 |
| chr1_-_167466780 | 1.56 |
ENSMUST00000036643.4
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr5_-_44099220 | 1.55 |
ENSMUST00000165909.1
|
Prom1
|
prominin 1 |
| chr11_-_100261021 | 1.53 |
ENSMUST00000080893.6
|
Krt17
|
keratin 17 |
| chr5_+_108369858 | 1.53 |
ENSMUST00000100944.2
|
Gm10419
|
predicted gene 10419 |
| chr4_-_126325672 | 1.51 |
ENSMUST00000102616.1
|
Tekt2
|
tektin 2 |
| chr9_+_124101944 | 1.47 |
ENSMUST00000171719.1
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr11_+_29463735 | 1.46 |
ENSMUST00000155854.1
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr7_-_134232125 | 1.44 |
ENSMUST00000127524.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr13_+_83504032 | 1.41 |
ENSMUST00000163888.1
ENSMUST00000005722.7 |
Mef2c
|
myocyte enhancer factor 2C |
| chr13_+_91461050 | 1.41 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr4_-_126325641 | 1.40 |
ENSMUST00000131113.1
|
Tekt2
|
tektin 2 |
| chr5_+_13399309 | 1.38 |
ENSMUST00000030714.7
ENSMUST00000141968.1 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_+_166344692 | 1.36 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chr3_-_106219477 | 1.35 |
ENSMUST00000082219.5
|
Chi3l4
|
chitinase 3-like 4 |
| chr6_-_82774448 | 1.35 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr11_+_44617310 | 1.31 |
ENSMUST00000081265.5
ENSMUST00000101326.3 ENSMUST00000109268.1 |
Ebf1
|
early B cell factor 1 |
| chr8_+_45885479 | 1.31 |
ENSMUST00000034053.5
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr11_-_79530569 | 1.31 |
ENSMUST00000103236.3
ENSMUST00000170799.1 ENSMUST00000170422.2 |
Evi2a
Evi2b
|
ecotropic viral integration site 2a ecotropic viral integration site 2b |
| chr18_+_64254359 | 1.30 |
ENSMUST00000025477.7
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr5_-_89883321 | 1.30 |
ENSMUST00000163159.1
ENSMUST00000061427.5 |
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3 |
| chr10_+_100488289 | 1.29 |
ENSMUST00000164751.1
|
Cep290
|
centrosomal protein 290 |
| chrX_-_51681856 | 1.29 |
ENSMUST00000114871.1
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr7_+_100494044 | 1.27 |
ENSMUST00000153287.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr15_+_78926720 | 1.25 |
ENSMUST00000089377.5
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr16_-_13986855 | 1.24 |
ENSMUST00000117803.1
|
Ifitm7
|
interferon induced transmembrane protein 7 |
| chrX_-_51681703 | 1.24 |
ENSMUST00000088172.5
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr13_-_100786402 | 1.23 |
ENSMUST00000174038.1
ENSMUST00000091295.7 ENSMUST00000072119.8 |
Ccnb1
|
cyclin B1 |
| chr1_-_95667555 | 1.23 |
ENSMUST00000043336.4
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr3_-_67463828 | 1.22 |
ENSMUST00000058981.2
|
Lxn
|
latexin |
| chr3_-_129755305 | 1.19 |
ENSMUST00000029653.2
|
Egf
|
epidermal growth factor |
| chr7_-_127137807 | 1.18 |
ENSMUST00000049931.5
|
Spn
|
sialophorin |
| chr7_+_100493795 | 1.16 |
ENSMUST00000129324.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr1_-_170927567 | 1.16 |
ENSMUST00000046322.7
ENSMUST00000159171.1 |
Fcrla
|
Fc receptor-like A |
| chr7_+_118712516 | 1.16 |
ENSMUST00000106557.1
|
Ccp110
|
centriolar coiled coil protein 110 |
| chr11_+_108682602 | 1.14 |
ENSMUST00000106718.3
ENSMUST00000106715.1 ENSMUST00000106724.3 |
Cep112
|
centrosomal protein 112 |
| chr9_-_60511003 | 1.13 |
ENSMUST00000098660.3
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr19_-_50678642 | 1.11 |
ENSMUST00000072685.6
ENSMUST00000164039.2 |
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr7_+_140218258 | 1.10 |
ENSMUST00000084460.6
|
Cd163l1
|
CD163 molecule-like 1 |
| chr5_-_138207302 | 1.09 |
ENSMUST00000160126.1
|
Gm454
|
predicted gene 454 |
| chr11_-_76027726 | 1.09 |
ENSMUST00000021207.6
|
Fam101b
|
family with sequence similarity 101, member B |
| chrX_+_164162167 | 1.08 |
ENSMUST00000131543.1
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr10_-_28986280 | 1.08 |
ENSMUST00000152363.1
ENSMUST00000015663.6 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr1_-_120505084 | 1.07 |
ENSMUST00000027639.1
|
Marco
|
macrophage receptor with collagenous structure |
| chr11_+_120361506 | 1.07 |
ENSMUST00000026445.2
|
Fscn2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr3_+_146121655 | 1.06 |
ENSMUST00000039450.4
|
Mcoln3
|
mucolipin 3 |
| chr4_-_116405986 | 1.05 |
ENSMUST00000123072.1
ENSMUST00000144281.1 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chrX_+_159627265 | 1.05 |
ENSMUST00000112456.2
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr1_+_134037490 | 1.05 |
ENSMUST00000162779.1
|
Fmod
|
fibromodulin |
| chr3_+_88532314 | 1.04 |
ENSMUST00000172699.1
|
Mex3a
|
mex3 homolog A (C. elegans) |
| chr15_-_56694525 | 1.04 |
ENSMUST00000050544.7
|
Has2
|
hyaluronan synthase 2 |
| chr9_-_121792478 | 1.00 |
ENSMUST00000035110.4
|
Hhatl
|
hedgehog acyltransferase-like |
| chr10_+_42860776 | 1.00 |
ENSMUST00000105494.1
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr13_-_117025505 | 1.00 |
ENSMUST00000022239.6
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_149309444 | 0.97 |
ENSMUST00000100765.4
|
2810474O19Rik
|
RIKEN cDNA 2810474O19 gene |
| chr16_-_3718105 | 0.95 |
ENSMUST00000023180.7
ENSMUST00000100222.2 |
Mefv
|
Mediterranean fever |
| chr11_+_9191934 | 0.94 |
ENSMUST00000042740.6
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr11_+_96282529 | 0.93 |
ENSMUST00000125410.1
|
Hoxb8
|
homeobox B8 |
| chr1_-_156204998 | 0.93 |
ENSMUST00000015628.3
|
Fam163a
|
family with sequence similarity 163, member A |
| chrX_-_164258186 | 0.93 |
ENSMUST00000112265.2
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr11_+_32000496 | 0.92 |
ENSMUST00000093219.3
|
Nsg2
|
neuron specific gene family member 2 |
| chr5_+_138229822 | 0.92 |
ENSMUST00000159798.1
ENSMUST00000159964.1 |
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr18_+_37020097 | 0.91 |
ENSMUST00000047614.1
|
Pcdha2
|
protocadherin alpha 2 |
| chr2_+_74704615 | 0.91 |
ENSMUST00000151380.1
|
Hoxd8
|
homeobox D8 |
| chr10_+_42860348 | 0.89 |
ENSMUST00000063063.7
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr1_-_170927540 | 0.88 |
ENSMUST00000162136.1
ENSMUST00000162887.1 |
Fcrla
|
Fc receptor-like A |
| chr6_-_28397999 | 0.87 |
ENSMUST00000035930.4
|
Zfp800
|
zinc finger protein 800 |
| chr8_+_84970068 | 0.87 |
ENSMUST00000164807.1
|
Prdx2
|
peroxiredoxin 2 |
| chr11_+_97415527 | 0.85 |
ENSMUST00000121799.1
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr6_+_149309391 | 0.85 |
ENSMUST00000130664.1
ENSMUST00000046689.6 |
2810474O19Rik
|
RIKEN cDNA 2810474O19 gene |
| chrX_+_72987339 | 0.84 |
ENSMUST00000164800.1
ENSMUST00000114546.2 |
Zfp185
|
zinc finger protein 185 |
| chr11_-_70656467 | 0.84 |
ENSMUST00000131642.1
|
Gm12319
|
predicted gene 12319 |
| chr7_+_81523531 | 0.84 |
ENSMUST00000181903.1
|
2900076A07Rik
|
RIKEN cDNA 2900076A07 gene |
| chr7_-_113347273 | 0.83 |
ENSMUST00000117577.1
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr10_+_127048235 | 0.81 |
ENSMUST00000165764.1
|
Cyp27b1
|
cytochrome P450, family 27, subfamily b, polypeptide 1 |
| chr4_+_132351768 | 0.81 |
ENSMUST00000172202.1
|
Gm17300
|
predicted gene, 17300 |
| chr18_-_13972617 | 0.81 |
ENSMUST00000025288.7
|
Zfp521
|
zinc finger protein 521 |
| chr7_-_102477902 | 0.81 |
ENSMUST00000061482.5
|
Olfr543
|
olfactory receptor 543 |
| chr3_+_95499273 | 0.81 |
ENSMUST00000015664.3
|
Ctsk
|
cathepsin K |
| chr15_+_6579841 | 0.81 |
ENSMUST00000090461.5
|
Fyb
|
FYN binding protein |
| chr2_-_140671400 | 0.81 |
ENSMUST00000056760.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr2_+_155751117 | 0.80 |
ENSMUST00000029140.5
ENSMUST00000132608.1 |
Procr
|
protein C receptor, endothelial |
| chr7_+_28071230 | 0.80 |
ENSMUST00000138392.1
ENSMUST00000076648.7 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr11_+_63133068 | 0.80 |
ENSMUST00000108700.1
|
Pmp22
|
peripheral myelin protein 22 |
| chr7_+_75643223 | 0.79 |
ENSMUST00000137959.1
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr18_-_54990124 | 0.79 |
ENSMUST00000064763.5
|
Zfp608
|
zinc finger protein 608 |
| chr4_-_133872304 | 0.78 |
ENSMUST00000157067.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chrX_+_123581700 | 0.78 |
ENSMUST00000178457.1
|
Gm6604
|
predicted gene 6604 |
| chr14_+_52016849 | 0.77 |
ENSMUST00000100638.2
|
Tmem253
|
transmembrane protein 253 |
| chr11_-_53618659 | 0.76 |
ENSMUST00000000889.6
|
Il4
|
interleukin 4 |
| chr11_+_44400060 | 0.75 |
ENSMUST00000102796.3
ENSMUST00000170513.1 |
Il12b
|
interleukin 12b |
| chr8_+_106510853 | 0.75 |
ENSMUST00000080797.6
|
Cdh3
|
cadherin 3 |
| chr3_-_152266320 | 0.75 |
ENSMUST00000046045.8
|
Nexn
|
nexilin |
| chr10_-_44458715 | 0.74 |
ENSMUST00000039174.4
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr5_+_3543812 | 0.74 |
ENSMUST00000115527.3
|
Fam133b
|
family with sequence similarity 133, member B |
| chr1_-_23909687 | 0.74 |
ENSMUST00000129254.1
|
Smap1
|
small ArfGAP 1 |
| chr1_-_60043087 | 0.73 |
ENSMUST00000027172.6
|
Ica1l
|
islet cell autoantigen 1-like |
| chr11_+_63132569 | 0.73 |
ENSMUST00000108701.1
|
Pmp22
|
peripheral myelin protein 22 |
| chr9_+_64385626 | 0.72 |
ENSMUST00000093829.2
ENSMUST00000118485.1 ENSMUST00000164113.1 |
Megf11
|
multiple EGF-like-domains 11 |
| chr12_+_91400990 | 0.71 |
ENSMUST00000021346.7
ENSMUST00000021343.6 |
Tshr
|
thyroid stimulating hormone receptor |
| chr15_-_83251720 | 0.71 |
ENSMUST00000164614.1
ENSMUST00000049530.6 |
A4galt
|
alpha 1,4-galactosyltransferase |
| chr4_-_132351636 | 0.71 |
ENSMUST00000105951.1
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr11_-_99493112 | 0.71 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr18_-_38250157 | 0.70 |
ENSMUST00000181871.1
|
1700086O06Rik
|
RIKEN cDNA 1700086O06 gene |
| chr4_-_129558355 | 0.68 |
ENSMUST00000167288.1
ENSMUST00000134336.1 |
Lck
|
lymphocyte protein tyrosine kinase |
| chrX_-_105929397 | 0.68 |
ENSMUST00000113573.1
ENSMUST00000130980.1 |
Atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr19_+_10525244 | 0.68 |
ENSMUST00000038379.3
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr14_+_79515618 | 0.68 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr4_-_129558387 | 0.68 |
ENSMUST00000067240.4
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr11_+_67200052 | 0.68 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr1_-_91459254 | 0.67 |
ENSMUST00000069620.8
|
Per2
|
period circadian clock 2 |
| chrX_+_7822289 | 0.67 |
ENSMUST00000009875.4
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr2_-_121271315 | 0.67 |
ENSMUST00000131245.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr10_+_67185730 | 0.67 |
ENSMUST00000173689.1
|
Jmjd1c
|
jumonji domain containing 1C |
| chr8_+_19682268 | 0.66 |
ENSMUST00000153710.1
ENSMUST00000127799.1 |
Gm6483
|
predicted gene 6483 |
| chrX_+_103422010 | 0.65 |
ENSMUST00000182089.1
|
Gm26992
|
predicted gene, 26992 |
| chr4_-_11386679 | 0.65 |
ENSMUST00000043781.7
ENSMUST00000108310.1 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr3_+_14533788 | 0.64 |
ENSMUST00000108370.2
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr2_-_73529725 | 0.64 |
ENSMUST00000094681.4
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr8_-_69791170 | 0.64 |
ENSMUST00000131784.1
|
Zfp866
|
zinc finger protein 866 |
| chr15_+_61985540 | 0.64 |
ENSMUST00000159327.1
ENSMUST00000167731.1 |
Myc
|
myelocytomatosis oncogene |
| chr13_+_93304940 | 0.64 |
ENSMUST00000109497.1
ENSMUST00000109498.1 ENSMUST00000060490.4 ENSMUST00000109492.1 ENSMUST00000109496.1 ENSMUST00000109495.1 |
Homer1
|
homer homolog 1 (Drosophila) |
| chr10_-_62507737 | 0.63 |
ENSMUST00000020271.6
|
Srgn
|
serglycin |
| chr10_-_44458687 | 0.63 |
ENSMUST00000105490.2
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr1_+_21272338 | 0.63 |
ENSMUST00000178379.1
|
Gm7094
|
predicted pseudogene 7094 |
| chr2_-_124222331 | 0.63 |
ENSMUST00000136658.1
|
4930583P06Rik
|
RIKEN cDNA 4930583P06 gene |
| chr7_+_81523555 | 0.62 |
ENSMUST00000180385.1
ENSMUST00000180879.1 ENSMUST00000181164.1 ENSMUST00000181264.1 |
2900076A07Rik
|
RIKEN cDNA 2900076A07 gene |
| chr3_-_61365951 | 0.62 |
ENSMUST00000066298.2
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr19_-_5845471 | 0.62 |
ENSMUST00000174287.1
ENSMUST00000173672.1 |
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr3_+_14533817 | 0.62 |
ENSMUST00000169079.1
ENSMUST00000091325.3 |
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr12_+_58211772 | 0.61 |
ENSMUST00000110671.2
ENSMUST00000044299.2 |
Sstr1
|
somatostatin receptor 1 |
| chr18_-_10181792 | 0.61 |
ENSMUST00000067947.5
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr5_+_35893319 | 0.61 |
ENSMUST00000064571.4
|
Afap1
|
actin filament associated protein 1 |
| chr15_-_100424208 | 0.61 |
ENSMUST00000154331.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr13_+_44731281 | 0.59 |
ENSMUST00000174086.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr17_+_78508063 | 0.59 |
ENSMUST00000024880.9
|
Vit
|
vitrin |
| chr4_+_130047840 | 0.59 |
ENSMUST00000044565.8
ENSMUST00000132251.1 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr8_-_104624266 | 0.58 |
ENSMUST00000163783.2
|
Cdh16
|
cadherin 16 |
| chr14_+_53795455 | 0.58 |
ENSMUST00000103671.2
|
B230359F08Rik
|
RIKEN cDNA B230359F08 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:1990535 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 1.5 | 4.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.2 | 1.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 1.1 | 3.3 | GO:2000458 | immune complex clearance by monocytes and macrophages(GO:0002436) astrocyte chemotaxis(GO:0035700) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.8 | 2.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.8 | 4.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.7 | 2.2 | GO:0002879 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.7 | 3.0 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.7 | 0.7 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.6 | 1.9 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 0.6 | 5.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 1.6 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.5 | 1.5 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 1.4 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.5 | 1.4 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.4 | 2.6 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.4 | 2.6 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.4 | 2.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 1.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.4 | 1.2 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 0.4 | 2.6 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 1.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 1.3 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.3 | 1.5 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.3 | 1.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 2.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 | 1.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 1.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.3 | 1.4 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 2.7 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.3 | 1.1 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.3 | 0.8 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.3 | 1.0 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.2 | 0.9 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.2 | 0.5 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.2 | 0.7 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.2 | 1.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 2.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 1.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 1.6 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.2 | 0.6 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.2 | 2.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.8 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.2 | 1.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 | 0.7 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.2 | 1.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 2.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.6 | GO:0090096 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 | 0.8 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 | 0.6 | GO:0033380 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 2.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 0.6 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 6.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 2.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 2.0 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 2.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.5 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 3.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.7 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.1 | 0.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 1.4 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.9 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 2.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 1.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 0.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.2 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) |
| 0.1 | 0.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 1.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 1.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.2 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.2 | GO:0061235 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.3 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.5 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 4.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.5 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.5 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.8 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.7 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.6 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 1.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0019660 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 1.3 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.7 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 1.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.8 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 2.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.9 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.4 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 1.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 1.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.3 | 1.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 1.1 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.3 | 4.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 5.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 2.6 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 2.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 2.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 3.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 2.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 2.8 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.5 | 4.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.1 | 3.3 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.6 | 2.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.6 | 5.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 1.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 2.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 2.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 2.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.5 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.3 | 2.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.3 | 1.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.3 | 1.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 2.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 0.8 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.3 | 6.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 1.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.4 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 4.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 2.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 6.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.4 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 3.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 6.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 6.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 5.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 3.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 5.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 1.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 1.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 5.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 4.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 4.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |