Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
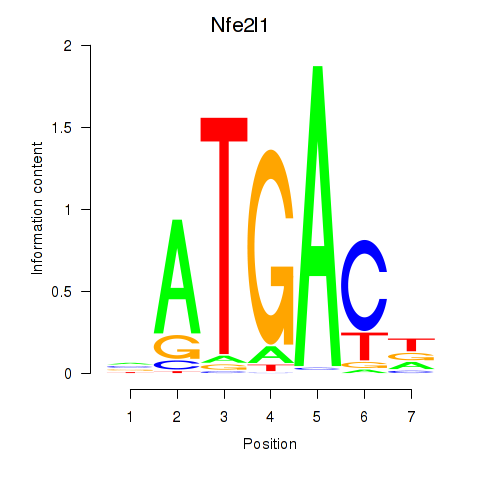
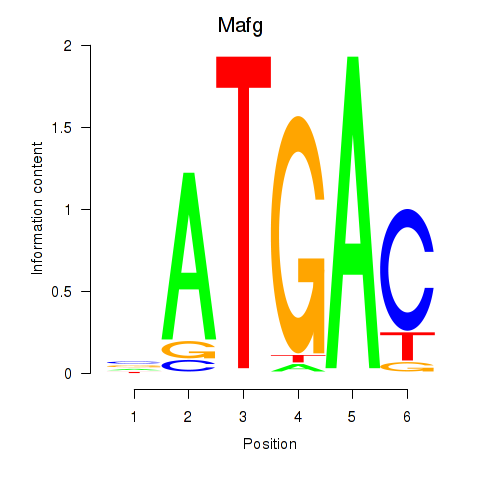
Results for Nfe2l1_Mafg
Z-value: 0.68
Transcription factors associated with Nfe2l1_Mafg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l1
|
ENSMUSG00000038615.11 | nuclear factor, erythroid derived 2,-like 1 |
|
Mafg
|
ENSMUSG00000051510.7 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
|
Mafg
|
ENSMUSG00000053906.4 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafg | mm10_v2_chr11_-_120630126_120630160 | 0.56 | 3.5e-04 | Click! |
| Nfe2l1 | mm10_v2_chr11_-_96829483_96829502 | -0.41 | 1.2e-02 | Click! |
Activity profile of Nfe2l1_Mafg motif
Sorted Z-values of Nfe2l1_Mafg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_49788204 | 3.75 |
ENSMUST00000114893.1
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr1_+_174158722 | 3.40 |
ENSMUST00000068403.3
|
Olfr420
|
olfactory receptor 420 |
| chr5_-_107726017 | 3.07 |
ENSMUST00000159263.2
|
Gfi1
|
growth factor independent 1 |
| chr1_-_133753681 | 2.49 |
ENSMUST00000125659.1
ENSMUST00000165602.2 ENSMUST00000048953.7 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr1_+_174041933 | 2.49 |
ENSMUST00000052975.4
|
Olfr433
|
olfactory receptor 433 |
| chr3_+_157566868 | 2.36 |
ENSMUST00000041175.6
ENSMUST00000173533.1 |
Ptger3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr1_-_82768449 | 2.27 |
ENSMUST00000027331.2
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr17_+_70929006 | 2.14 |
ENSMUST00000092011.6
|
Gm16519
|
predicted gene, 16519 |
| chr7_-_116038734 | 2.09 |
ENSMUST00000166877.1
|
Sox6
|
SRY-box containing gene 6 |
| chr1_-_132367879 | 1.94 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_18998126 | 1.90 |
ENSMUST00000006912.5
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr10_-_76110956 | 1.70 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr17_+_78491549 | 1.69 |
ENSMUST00000079363.4
|
Gm10093
|
predicted pseudogene 10093 |
| chr14_+_74640840 | 1.67 |
ENSMUST00000036653.3
|
Htr2a
|
5-hydroxytryptamine (serotonin) receptor 2A |
| chr6_-_129507107 | 1.58 |
ENSMUST00000183258.1
ENSMUST00000182784.1 ENSMUST00000032265.6 ENSMUST00000162815.1 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr1_-_45890078 | 1.58 |
ENSMUST00000183590.1
|
Gm5269
|
predicted gene 5269 |
| chr10_+_82985473 | 1.52 |
ENSMUST00000040110.7
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr2_+_18998332 | 1.50 |
ENSMUST00000028069.1
|
4930426L09Rik
|
RIKEN cDNA 4930426L09 gene |
| chr2_+_130277157 | 1.50 |
ENSMUST00000028890.8
ENSMUST00000159373.1 |
Nop56
|
NOP56 ribonucleoprotein |
| chr1_-_128328311 | 1.50 |
ENSMUST00000073490.6
|
Lct
|
lactase |
| chr11_+_95010277 | 1.49 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr8_+_123332676 | 1.48 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr17_-_48432723 | 1.44 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr2_-_28699636 | 1.38 |
ENSMUST00000102877.1
|
1700026L06Rik
|
RIKEN cDNA 1700026L06 gene |
| chr15_-_101694299 | 1.26 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr3_+_127553462 | 1.20 |
ENSMUST00000043108.4
|
4930422G04Rik
|
RIKEN cDNA 4930422G04 gene |
| chr15_-_101924725 | 1.14 |
ENSMUST00000023797.6
|
Krt4
|
keratin 4 |
| chr12_-_54986363 | 1.12 |
ENSMUST00000173433.1
ENSMUST00000173803.1 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr1_+_172956768 | 1.11 |
ENSMUST00000038432.4
|
Olfr16
|
olfactory receptor 16 |
| chr10_-_79874233 | 1.09 |
ENSMUST00000166023.1
ENSMUST00000167707.1 ENSMUST00000165601.1 |
BC005764
|
cDNA sequence BC005764 |
| chr10_-_79874211 | 1.08 |
ENSMUST00000167897.1
|
BC005764
|
cDNA sequence BC005764 |
| chr13_-_3804307 | 1.05 |
ENSMUST00000077698.3
|
Calml3
|
calmodulin-like 3 |
| chr7_-_102477902 | 1.05 |
ENSMUST00000061482.5
|
Olfr543
|
olfactory receptor 543 |
| chr14_-_76237353 | 1.02 |
ENSMUST00000095471.4
|
Rps2-ps6
|
ribosomal protein S2, pseudogene 6 |
| chr6_-_86669136 | 0.98 |
ENSMUST00000001184.7
|
Mxd1
|
MAX dimerization protein 1 |
| chr17_+_35821675 | 0.98 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr18_-_9282754 | 0.97 |
ENSMUST00000041007.3
|
Gjd4
|
gap junction protein, delta 4 |
| chrX_+_56454871 | 0.93 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr10_+_52022502 | 0.93 |
ENSMUST00000163017.1
ENSMUST00000058347.4 |
Vgll2
|
vestigial like 2 homolog (Drosophila) |
| chr15_-_101491509 | 0.92 |
ENSMUST00000023718.7
|
5430421N21Rik
|
RIKEN cDNA 5430421N21 gene |
| chr17_-_40880525 | 0.87 |
ENSMUST00000068258.2
|
9130008F23Rik
|
RIKEN cDNA 9130008F23 gene |
| chr2_-_121271403 | 0.87 |
ENSMUST00000110648.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr8_+_94984399 | 0.86 |
ENSMUST00000093271.6
|
Gpr56
|
G protein-coupled receptor 56 |
| chr18_-_62179948 | 0.86 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr4_-_43558386 | 0.85 |
ENSMUST00000130353.1
|
Tln1
|
talin 1 |
| chr12_+_17348422 | 0.84 |
ENSMUST00000046011.10
|
Nol10
|
nucleolar protein 10 |
| chr2_-_121271341 | 0.83 |
ENSMUST00000110647.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr1_+_82839449 | 0.83 |
ENSMUST00000113444.1
ENSMUST00000063380.4 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr10_+_76147451 | 0.83 |
ENSMUST00000020450.3
|
Slc5a4a
|
solute carrier family 5, member 4a |
| chr13_+_53525703 | 0.82 |
ENSMUST00000081132.4
|
Gm5449
|
predicted pseudogene 5449 |
| chr9_+_96196246 | 0.81 |
ENSMUST00000165120.2
ENSMUST00000034982.9 |
Tfdp2
|
transcription factor Dp 2 |
| chr14_-_57826128 | 0.81 |
ENSMUST00000022536.2
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr4_-_117125618 | 0.78 |
ENSMUST00000183310.1
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr3_-_103791075 | 0.76 |
ENSMUST00000106845.2
ENSMUST00000029438.8 ENSMUST00000121324.1 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr9_+_108560422 | 0.74 |
ENSMUST00000081111.8
|
Impdh2
|
inosine 5'-phosphate dehydrogenase 2 |
| chr1_+_135729147 | 0.72 |
ENSMUST00000027677.7
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chrX_-_75130996 | 0.71 |
ENSMUST00000033775.2
|
Mpp1
|
membrane protein, palmitoylated |
| chr4_+_101507855 | 0.70 |
ENSMUST00000038207.5
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr9_+_108508005 | 0.70 |
ENSMUST00000006838.8
ENSMUST00000134939.1 |
Qars
|
glutaminyl-tRNA synthetase |
| chr15_+_35296090 | 0.68 |
ENSMUST00000022952.4
|
Osr2
|
odd-skipped related 2 |
| chr4_-_63403330 | 0.68 |
ENSMUST00000035724.4
|
Akna
|
AT-hook transcription factor |
| chr3_-_103791537 | 0.66 |
ENSMUST00000118317.1
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr3_+_88629442 | 0.66 |
ENSMUST00000176316.1
ENSMUST00000176879.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_12846773 | 0.66 |
ENSMUST00000164001.1
|
Gm5244
|
predicted pseudogene 5244 |
| chr11_+_48800357 | 0.65 |
ENSMUST00000020640.7
|
Gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 like 1 |
| chr6_-_136835426 | 0.65 |
ENSMUST00000068293.3
ENSMUST00000111894.1 |
Smco3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr11_+_44617310 | 0.64 |
ENSMUST00000081265.5
ENSMUST00000101326.3 ENSMUST00000109268.1 |
Ebf1
|
early B cell factor 1 |
| chr7_+_30776394 | 0.64 |
ENSMUST00000041703.7
|
Dmkn
|
dermokine |
| chrX_-_139714481 | 0.63 |
ENSMUST00000183728.1
|
Gm15013
|
predicted gene 15013 |
| chr2_-_121271315 | 0.62 |
ENSMUST00000131245.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr10_-_84533884 | 0.62 |
ENSMUST00000053871.3
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr7_-_16917184 | 0.60 |
ENSMUST00000173139.1
|
Calm3
|
calmodulin 3 |
| chrX_-_75130914 | 0.60 |
ENSMUST00000114091.1
|
Mpp1
|
membrane protein, palmitoylated |
| chr4_-_132049058 | 0.60 |
ENSMUST00000105981.2
ENSMUST00000084253.3 ENSMUST00000141291.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr11_+_78826575 | 0.60 |
ENSMUST00000147875.2
ENSMUST00000141321.1 |
Lyrm9
|
LYR motif containing 9 |
| chr14_+_11227511 | 0.60 |
ENSMUST00000080237.3
|
Rpl21-ps4
|
ribosomal protein L21, pseudogene 4 |
| chr14_+_53709902 | 0.58 |
ENSMUST00000103638.4
|
Trav6-7-dv9
|
T cell receptor alpha variable 6-7-DV9 |
| chr10_+_23949516 | 0.58 |
ENSMUST00000045152.4
|
Taar3
|
trace amine-associated receptor 3 |
| chrX_-_75130844 | 0.57 |
ENSMUST00000114092.1
ENSMUST00000132501.1 ENSMUST00000153318.1 ENSMUST00000155742.1 |
Mpp1
|
membrane protein, palmitoylated |
| chr6_-_99044414 | 0.57 |
ENSMUST00000177507.1
ENSMUST00000123992.1 |
Foxp1
|
forkhead box P1 |
| chr10_-_84533968 | 0.57 |
ENSMUST00000167671.1
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr5_+_35057059 | 0.56 |
ENSMUST00000050709.3
|
Dok7
|
docking protein 7 |
| chr1_+_127774164 | 0.55 |
ENSMUST00000027587.8
ENSMUST00000112570.1 |
Ccnt2
|
cyclin T2 |
| chr3_+_88616133 | 0.55 |
ENSMUST00000176500.1
ENSMUST00000177498.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr17_-_55915870 | 0.54 |
ENSMUST00000074828.4
|
Rpl21-ps6
|
ribosomal protein L21, pseudogene 6 |
| chr13_-_55528511 | 0.54 |
ENSMUST00000047877.4
|
Dok3
|
docking protein 3 |
| chr11_-_33276334 | 0.53 |
ENSMUST00000183831.1
|
Gm12117
|
predicted gene 12117 |
| chr5_-_65391380 | 0.53 |
ENSMUST00000120094.1
ENSMUST00000118543.1 ENSMUST00000127874.1 |
Rpl9
|
ribosomal protein L9 |
| chr5_-_65391408 | 0.51 |
ENSMUST00000057885.6
|
Rpl9
|
ribosomal protein L9 |
| chr3_+_88629499 | 0.51 |
ENSMUST00000175745.1
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr11_-_103208542 | 0.51 |
ENSMUST00000021323.4
ENSMUST00000107026.2 |
1700023F06Rik
|
RIKEN cDNA 1700023F06 gene |
| chr18_-_15063560 | 0.49 |
ENSMUST00000168989.1
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr11_-_59964936 | 0.49 |
ENSMUST00000062405.7
|
Rasd1
|
RAS, dexamethasone-induced 1 |
| chr13_-_32338565 | 0.47 |
ENSMUST00000041859.7
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr11_-_83649349 | 0.46 |
ENSMUST00000001008.5
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr9_+_48450327 | 0.46 |
ENSMUST00000165252.1
|
Gm5616
|
predicted gene 5616 |
| chr11_-_61719946 | 0.46 |
ENSMUST00000151780.1
ENSMUST00000148584.1 |
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr15_+_79690869 | 0.46 |
ENSMUST00000046463.8
|
Gtpbp1
|
GTP binding protein 1 |
| chr6_+_36388055 | 0.46 |
ENSMUST00000172278.1
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr5_+_31698050 | 0.45 |
ENSMUST00000114507.3
ENSMUST00000063813.4 ENSMUST00000071531.5 ENSMUST00000131995.1 |
Bre
|
brain and reproductive organ-expressed protein |
| chr8_-_124021008 | 0.43 |
ENSMUST00000093039.5
|
Taf5l
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr17_-_44735612 | 0.42 |
ENSMUST00000162373.1
ENSMUST00000162878.1 |
Runx2
|
runt related transcription factor 2 |
| chr12_-_10900296 | 0.42 |
ENSMUST00000085735.2
|
Pgk1-rs7
|
phosphoglycerate kinase-1, related sequence-7 |
| chr1_-_86388162 | 0.41 |
ENSMUST00000027440.3
|
Nmur1
|
neuromedin U receptor 1 |
| chr10_-_95673451 | 0.41 |
ENSMUST00000099328.1
|
Anapc15-ps
|
anaphase prompoting complex C subunit 15, pseudogene |
| chr7_+_103928825 | 0.41 |
ENSMUST00000106863.1
|
Olfr631
|
olfactory receptor 631 |
| chr17_+_7925990 | 0.40 |
ENSMUST00000036370.7
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr3_+_130068390 | 0.40 |
ENSMUST00000076703.6
|
Gm9396
|
predicted gene 9396 |
| chr13_-_68999518 | 0.40 |
ENSMUST00000022013.7
|
Adcy2
|
adenylate cyclase 2 |
| chr18_-_10030017 | 0.40 |
ENSMUST00000116669.1
ENSMUST00000092096.6 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr6_+_40964760 | 0.39 |
ENSMUST00000076638.5
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr7_+_101896817 | 0.38 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr14_+_79481164 | 0.38 |
ENSMUST00000040131.5
|
Elf1
|
E74-like factor 1 |
| chr7_+_104003259 | 0.38 |
ENSMUST00000098184.1
|
Olfr638
|
olfactory receptor 638 |
| chr10_+_80930071 | 0.37 |
ENSMUST00000015456.8
|
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr6_+_128438757 | 0.37 |
ENSMUST00000144745.1
|
Gm10069
|
predicted gene 10069 |
| chr1_-_43163891 | 0.37 |
ENSMUST00000008280.7
|
Fhl2
|
four and a half LIM domains 2 |
| chr5_+_31697960 | 0.37 |
ENSMUST00000114515.2
|
Bre
|
brain and reproductive organ-expressed protein |
| chr14_+_54625305 | 0.37 |
ENSMUST00000097177.4
|
Psmb11
|
proteasome (prosome, macropain) subunit, beta type, 11 |
| chr2_-_7395879 | 0.37 |
ENSMUST00000182404.1
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr18_+_6332587 | 0.36 |
ENSMUST00000097682.2
|
Rpl27-ps3
|
ribosomal protein L27, pseudogene 3 |
| chr2_-_7395968 | 0.35 |
ENSMUST00000002176.6
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr15_-_42676967 | 0.35 |
ENSMUST00000022921.5
|
Angpt1
|
angiopoietin 1 |
| chr10_-_86732409 | 0.35 |
ENSMUST00000070435.4
|
Fabp3-ps1
|
fatty acid binding protein 3, muscle and heart, pseudogene 1 |
| chr10_+_111164794 | 0.35 |
ENSMUST00000105275.1
ENSMUST00000095310.1 |
Osbpl8
|
oxysterol binding protein-like 8 |
| chr11_+_51619731 | 0.34 |
ENSMUST00000127405.1
|
Nhp2
|
NHP2 ribonucleoprotein |
| chrX_+_71556874 | 0.34 |
ENSMUST00000123100.1
|
Hmgb3
|
high mobility group box 3 |
| chr8_+_94810446 | 0.34 |
ENSMUST00000034232.1
|
Ccl17
|
chemokine (C-C motif) ligand 17 |
| chr7_+_66365905 | 0.33 |
ENSMUST00000107486.1
|
Gm10974
|
predicted gene 10974 |
| chr14_+_50392758 | 0.32 |
ENSMUST00000058965.3
|
Olfr736
|
olfactory receptor 736 |
| chr2_-_30903255 | 0.32 |
ENSMUST00000102852.3
|
Ptges
|
prostaglandin E synthase |
| chr1_-_183147461 | 0.32 |
ENSMUST00000171366.1
|
Disp1
|
dispatched homolog 1 (Drosophila) |
| chr11_+_115933282 | 0.32 |
ENSMUST00000140991.1
|
Sap30bp
|
SAP30 binding protein |
| chr11_-_52000748 | 0.32 |
ENSMUST00000109086.1
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr19_-_60874526 | 0.31 |
ENSMUST00000025961.6
|
Prdx3
|
peroxiredoxin 3 |
| chr14_-_79481268 | 0.31 |
ENSMUST00000022601.5
|
Wbp4
|
WW domain binding protein 4 |
| chr18_-_47333311 | 0.29 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr9_+_53771499 | 0.29 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chrX_+_20688379 | 0.29 |
ENSMUST00000033380.6
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr1_-_191183244 | 0.29 |
ENSMUST00000027941.8
|
Atf3
|
activating transcription factor 3 |
| chr6_-_28830345 | 0.29 |
ENSMUST00000171353.1
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr8_-_40511663 | 0.27 |
ENSMUST00000135269.1
ENSMUST00000034012.3 |
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr5_+_134676490 | 0.27 |
ENSMUST00000100641.2
|
Gm10369
|
predicted gene 10369 |
| chr10_+_13008442 | 0.27 |
ENSMUST00000105139.3
|
Sf3b5
|
splicing factor 3b, subunit 5 |
| chr7_-_84679346 | 0.27 |
ENSMUST00000069537.2
ENSMUST00000178385.1 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr12_-_31634592 | 0.26 |
ENSMUST00000020979.7
ENSMUST00000177962.1 |
Bcap29
|
B cell receptor associated protein 29 |
| chr8_-_35826435 | 0.26 |
ENSMUST00000060128.5
|
Cldn23
|
claudin 23 |
| chr17_-_70998010 | 0.26 |
ENSMUST00000024846.6
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr7_-_103741322 | 0.26 |
ENSMUST00000051346.2
|
Olfr629
|
olfactory receptor 629 |
| chr8_-_40511298 | 0.25 |
ENSMUST00000149992.1
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr15_-_102189032 | 0.25 |
ENSMUST00000023805.1
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr8_-_60954726 | 0.25 |
ENSMUST00000110302.1
|
Clcn3
|
chloride channel 3 |
| chr9_-_123113158 | 0.24 |
ENSMUST00000147563.1
|
Zdhhc3
|
zinc finger, DHHC domain containing 3 |
| chrX_+_71663665 | 0.24 |
ENSMUST00000070449.5
|
Gpr50
|
G-protein-coupled receptor 50 |
| chr7_+_118633729 | 0.24 |
ENSMUST00000057320.7
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr1_+_87404916 | 0.24 |
ENSMUST00000173152.1
ENSMUST00000173663.1 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr6_+_92816460 | 0.24 |
ENSMUST00000057977.3
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chrX_-_141874870 | 0.24 |
ENSMUST00000182079.1
|
Gm15294
|
predicted gene 15294 |
| chr16_+_20694908 | 0.23 |
ENSMUST00000056518.6
|
Fam131a
|
family with sequence similarity 131, member A |
| chr19_-_5366285 | 0.23 |
ENSMUST00000170010.1
|
Banf1
|
barrier to autointegration factor 1 |
| chr18_-_62741387 | 0.23 |
ENSMUST00000097557.3
|
Spink13
|
serine peptidase inhibitor, Kazal type 13 |
| chr14_-_52213379 | 0.23 |
ENSMUST00000140603.1
|
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr2_-_165283599 | 0.23 |
ENSMUST00000155289.1
|
Slc35c2
|
solute carrier family 35, member C2 |
| chr19_+_38132767 | 0.23 |
ENSMUST00000025956.5
ENSMUST00000112329.1 |
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr18_-_43477764 | 0.23 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_-_116853083 | 0.23 |
ENSMUST00000092404.6
|
Srsf2
|
serine/arginine-rich splicing factor 2 |
| chr12_-_103338314 | 0.23 |
ENSMUST00000149431.1
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr2_+_130406478 | 0.23 |
ENSMUST00000055421.4
|
Tmem239
|
transmembrane 239 |
| chr10_-_53750880 | 0.22 |
ENSMUST00000020003.7
|
Fam184a
|
family with sequence similarity 184, member A |
| chr2_-_25272380 | 0.22 |
ENSMUST00000028342.6
|
Ssna1
|
Sjogren's syndrome nuclear autoantigen 1 |
| chr17_-_46282991 | 0.22 |
ENSMUST00000180283.1
ENSMUST00000012440.6 ENSMUST00000164342.2 |
Tjap1
|
tight junction associated protein 1 |
| chr11_-_48826500 | 0.22 |
ENSMUST00000161192.2
|
Gm12184
|
predicted gene 12184 |
| chr5_+_107497762 | 0.22 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr14_+_53806497 | 0.22 |
ENSMUST00000103672.4
|
Trav17
|
T cell receptor alpha variable 17 |
| chr14_-_121965128 | 0.22 |
ENSMUST00000049872.7
|
Gpr183
|
G protein-coupled receptor 183 |
| chr17_-_71459300 | 0.21 |
ENSMUST00000183937.1
|
Gm4707
|
predicted gene 4707 |
| chr19_-_11283813 | 0.21 |
ENSMUST00000067673.6
|
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr17_+_71204647 | 0.20 |
ENSMUST00000126681.1
|
Lpin2
|
lipin 2 |
| chr9_-_50344981 | 0.20 |
ENSMUST00000076364.4
|
Rpl10-ps3
|
ribosomal protein L10, pseudogene 3 |
| chr8_+_124897877 | 0.20 |
ENSMUST00000034467.5
|
Sprtn
|
SprT-like N-terminal domain |
| chr2_-_76870486 | 0.19 |
ENSMUST00000138542.1
|
Ttn
|
titin |
| chr11_-_120348091 | 0.19 |
ENSMUST00000106215.4
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr1_-_182158319 | 0.19 |
ENSMUST00000169123.2
|
Vmn1r1
|
vomeronasal 1 receptor 1 |
| chr9_-_106887000 | 0.19 |
ENSMUST00000055843.7
|
Rbm15b
|
RNA binding motif protein 15B |
| chr15_-_81104999 | 0.19 |
ENSMUST00000109579.2
|
Mkl1
|
MKL (megakaryoblastic leukemia)/myocardin-like 1 |
| chr4_+_127021311 | 0.19 |
ENSMUST00000030623.7
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr12_-_31950535 | 0.19 |
ENSMUST00000172314.2
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr8_+_12947935 | 0.19 |
ENSMUST00000110871.1
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr15_-_41869703 | 0.18 |
ENSMUST00000054742.5
|
Abra
|
actin-binding Rho activating protein |
| chr14_-_52036143 | 0.18 |
ENSMUST00000052560.4
|
Olfr221
|
olfactory receptor 221 |
| chr4_+_133176336 | 0.18 |
ENSMUST00000105912.1
|
Wasf2
|
WAS protein family, member 2 |
| chr3_-_92500493 | 0.18 |
ENSMUST00000062129.1
|
Sprr4
|
small proline-rich protein 4 |
| chr2_+_119742306 | 0.18 |
ENSMUST00000028758.7
|
Itpka
|
inositol 1,4,5-trisphosphate 3-kinase A |
| chr10_-_127288999 | 0.18 |
ENSMUST00000119078.1
|
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr3_-_37232565 | 0.17 |
ENSMUST00000161015.1
ENSMUST00000029273.1 |
Il21
|
interleukin 21 |
| chr11_-_52000432 | 0.17 |
ENSMUST00000020657.6
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr17_-_35643684 | 0.17 |
ENSMUST00000095467.3
|
Dpcr1
|
diffuse panbronchiolitis critical region 1 (human) |
| chr12_+_38783503 | 0.17 |
ENSMUST00000159334.1
|
Etv1
|
ets variant gene 1 |
| chr15_+_6579841 | 0.17 |
ENSMUST00000090461.5
|
Fyb
|
FYN binding protein |
| chr14_+_54894133 | 0.17 |
ENSMUST00000116476.2
ENSMUST00000022808.7 ENSMUST00000150975.1 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l1_Mafg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.8 | 2.4 | GO:1990764 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of actin filament-based movement(GO:1903116) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.6 | 3.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.6 | 1.7 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.5 | 1.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.3 | 1.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 1.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.2 | 0.9 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 1.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 3.0 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 2.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 1.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 0.6 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 0.7 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 2.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 0.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 1.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.7 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 1.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 0.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 5.6 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 1.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.2 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 0.2 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.1 | 0.3 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.2 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.6 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.5 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 1.3 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 | 0.5 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.0 | 1.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.8 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.5 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 1.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.1 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.4 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.4 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.7 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.7 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.0 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 1.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.5 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 0.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 2.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 3.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.5 | 1.5 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.4 | 3.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 2.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 2.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 1.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 1.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 0.9 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.2 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 3.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.2 | 2.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 1.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 6.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 6.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 2.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |