Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
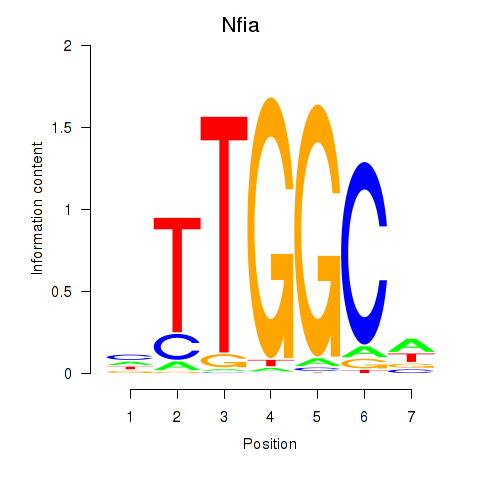
Results for Nfia
Z-value: 1.88
Transcription factors associated with Nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfia
|
ENSMUSG00000028565.12 | nuclear factor I/A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfia | mm10_v2_chr4_+_97777780_97777834 | 0.53 | 9.5e-04 | Click! |
Activity profile of Nfia motif
Sorted Z-values of Nfia motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_97417730 | 16.40 |
ENSMUST00000043077.7
|
Thrsp
|
thyroid hormone responsive |
| chr8_-_93131271 | 14.30 |
ENSMUST00000034189.8
|
Ces1c
|
carboxylesterase 1C |
| chr4_-_62054112 | 11.87 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr7_+_140763739 | 11.58 |
ENSMUST00000026552.7
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr1_+_167618246 | 10.28 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr3_-_81975742 | 10.03 |
ENSMUST00000029645.8
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr4_-_61674094 | 9.91 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_60421933 | 9.29 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_60582152 | 9.27 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr7_-_48848023 | 9.03 |
ENSMUST00000032658.6
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr3_+_94377432 | 8.88 |
ENSMUST00000107292.1
|
Rorc
|
RAR-related orphan receptor gamma |
| chr8_+_105131800 | 8.77 |
ENSMUST00000161289.1
|
Ces4a
|
carboxylesterase 4A |
| chr3_+_94377505 | 8.54 |
ENSMUST00000098877.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr11_-_5915124 | 8.44 |
ENSMUST00000109823.2
ENSMUST00000109822.1 |
Gck
|
glucokinase |
| chr6_-_141856171 | 8.16 |
ENSMUST00000165990.1
ENSMUST00000163678.1 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr4_-_60662358 | 8.11 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr1_-_162898665 | 8.04 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr7_+_140845562 | 7.80 |
ENSMUST00000035300.5
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr10_-_109010955 | 7.59 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr19_-_44407703 | 7.55 |
ENSMUST00000041331.2
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr7_+_25897620 | 7.48 |
ENSMUST00000072438.6
ENSMUST00000005477.5 |
Cyp2b10
|
cytochrome P450, family 2, subfamily b, polypeptide 10 |
| chr4_-_60139857 | 7.44 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr1_+_88070765 | 7.37 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr10_-_128960965 | 6.97 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr1_-_162898484 | 6.67 |
ENSMUST00000143123.1
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr15_+_10314102 | 6.35 |
ENSMUST00000127467.1
|
Prlr
|
prolactin receptor |
| chr6_-_5193757 | 6.08 |
ENSMUST00000177159.1
ENSMUST00000176945.1 |
Pon1
|
paraoxonase 1 |
| chr11_+_101367542 | 6.08 |
ENSMUST00000019469.2
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr6_-_5193817 | 5.79 |
ENSMUST00000002663.5
|
Pon1
|
paraoxonase 1 |
| chr19_-_42202150 | 5.59 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr19_+_44203265 | 5.58 |
ENSMUST00000026220.5
|
Scd3
|
stearoyl-coenzyme A desaturase 3 |
| chr9_-_57683644 | 5.56 |
ENSMUST00000034860.3
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2 |
| chr17_+_32685655 | 5.48 |
ENSMUST00000008801.6
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr4_-_61835185 | 5.42 |
ENSMUST00000082287.2
|
Mup5
|
major urinary protein 5 |
| chr4_-_62150810 | 5.29 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr1_-_130661584 | 5.28 |
ENSMUST00000137276.2
|
C4bp
|
complement component 4 binding protein |
| chr1_-_130661613 | 5.25 |
ENSMUST00000027657.7
|
C4bp
|
complement component 4 binding protein |
| chr3_+_138277489 | 5.25 |
ENSMUST00000004232.9
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr16_+_44173271 | 5.12 |
ENSMUST00000088356.4
ENSMUST00000169582.1 |
Gm608
|
predicted gene 608 |
| chr1_+_88095054 | 5.11 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr15_-_3583146 | 5.04 |
ENSMUST00000110698.2
|
Ghr
|
growth hormone receptor |
| chr7_-_99695628 | 4.97 |
ENSMUST00000145381.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr3_+_130617645 | 4.95 |
ENSMUST00000163620.1
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr19_-_46672883 | 4.90 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chrX_-_143933089 | 4.81 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr1_+_87594545 | 4.76 |
ENSMUST00000165109.1
ENSMUST00000070898.5 |
Neu2
|
neuraminidase 2 |
| chr3_-_129332713 | 4.75 |
ENSMUST00000029658.7
|
Enpep
|
glutamyl aminopeptidase |
| chr3_+_94372794 | 4.69 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr16_+_91269759 | 4.67 |
ENSMUST00000056882.5
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr4_+_60003438 | 4.67 |
ENSMUST00000107517.1
ENSMUST00000107520.1 |
Mup6
|
major urinary protein 6 |
| chr7_+_130936172 | 4.65 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr2_+_173153048 | 4.61 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr10_-_24712133 | 4.54 |
ENSMUST00000105520.1
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr7_+_51878967 | 4.52 |
ENSMUST00000051912.6
|
Gas2
|
growth arrest specific 2 |
| chr2_+_122147680 | 4.51 |
ENSMUST00000102476.4
|
B2m
|
beta-2 microglobulin |
| chr7_-_99695572 | 4.50 |
ENSMUST00000137914.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr10_-_24711987 | 4.42 |
ENSMUST00000135846.1
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr11_-_84167466 | 4.41 |
ENSMUST00000050771.7
|
Gm11437
|
predicted gene 11437 |
| chr10_-_24101951 | 4.39 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr17_+_36942910 | 4.32 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr11_-_70514608 | 4.25 |
ENSMUST00000021179.3
|
Vmo1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr3_+_130617448 | 4.24 |
ENSMUST00000166187.1
ENSMUST00000072271.6 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr14_-_118052235 | 4.20 |
ENSMUST00000022725.2
|
Dct
|
dopachrome tautomerase |
| chr15_-_3583191 | 4.12 |
ENSMUST00000069451.4
|
Ghr
|
growth hormone receptor |
| chr5_-_77115145 | 4.12 |
ENSMUST00000081964.5
|
Hopx
|
HOP homeobox |
| chr3_-_90514250 | 4.10 |
ENSMUST00000107340.1
ENSMUST00000060738.8 |
S100a1
|
S100 calcium binding protein A1 |
| chr2_-_25470031 | 3.99 |
ENSMUST00000114251.1
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr7_-_105600103 | 3.98 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr14_+_65970610 | 3.88 |
ENSMUST00000127387.1
|
Clu
|
clusterin |
| chr11_+_48837465 | 3.87 |
ENSMUST00000046903.5
|
Trim7
|
tripartite motif-containing 7 |
| chr17_+_32685610 | 3.75 |
ENSMUST00000168171.1
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr11_-_4118778 | 3.75 |
ENSMUST00000003681.7
|
Sec14l2
|
SEC14-like 2 (S. cerevisiae) |
| chr1_+_74332596 | 3.74 |
ENSMUST00000087225.5
|
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr11_+_115462464 | 3.72 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr3_-_131303144 | 3.69 |
ENSMUST00000106337.2
|
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr2_+_164562579 | 3.64 |
ENSMUST00000017867.3
ENSMUST00000109344.2 ENSMUST00000109345.2 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr7_+_51879041 | 3.63 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chr10_+_75893398 | 3.59 |
ENSMUST00000009236.4
|
Derl3
|
Der1-like domain family, member 3 |
| chr2_-_25469742 | 3.57 |
ENSMUST00000114259.2
ENSMUST00000015234.6 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr14_+_65970804 | 3.56 |
ENSMUST00000138191.1
|
Clu
|
clusterin |
| chr3_+_97628804 | 3.54 |
ENSMUST00000107050.1
ENSMUST00000029729.8 ENSMUST00000107049.1 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr16_+_41532851 | 3.54 |
ENSMUST00000078873.4
|
Lsamp
|
limbic system-associated membrane protein |
| chr2_-_164638789 | 3.51 |
ENSMUST00000109336.1
|
Wfdc16
|
WAP four-disulfide core domain 16 |
| chr13_-_56296551 | 3.49 |
ENSMUST00000021970.9
|
Cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr4_+_43631935 | 3.47 |
ENSMUST00000030191.8
|
Npr2
|
natriuretic peptide receptor 2 |
| chr18_+_20665250 | 3.45 |
ENSMUST00000075312.3
|
Ttr
|
transthyretin |
| chr6_+_138140298 | 3.45 |
ENSMUST00000008684.4
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr19_+_44333092 | 3.45 |
ENSMUST00000058856.8
|
Scd4
|
stearoyl-coenzyme A desaturase 4 |
| chr17_-_34000257 | 3.44 |
ENSMUST00000087189.6
ENSMUST00000173075.1 ENSMUST00000172760.1 ENSMUST00000172912.1 ENSMUST00000025181.10 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr7_-_4630473 | 3.36 |
ENSMUST00000055085.6
|
Tmem86b
|
transmembrane protein 86B |
| chr19_+_42036025 | 3.36 |
ENSMUST00000026172.2
|
Ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr10_+_4611971 | 3.32 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr16_-_10543028 | 3.31 |
ENSMUST00000184863.1
ENSMUST00000038281.5 |
Dexi
|
dexamethasone-induced transcript |
| chr11_-_78422217 | 3.31 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr2_+_163820832 | 3.26 |
ENSMUST00000029188.7
|
Wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr14_+_21052574 | 3.26 |
ENSMUST00000045376.9
|
Adk
|
adenosine kinase |
| chr13_+_24943144 | 3.23 |
ENSMUST00000021773.5
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr2_+_85975213 | 3.21 |
ENSMUST00000082191.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr1_+_88055467 | 3.17 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr18_-_61911783 | 3.16 |
ENSMUST00000049378.8
ENSMUST00000166783.1 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chrX_-_100594860 | 3.11 |
ENSMUST00000053373.1
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr6_-_59024470 | 3.10 |
ENSMUST00000089860.5
|
Fam13a
|
family with sequence similarity 13, member A |
| chr17_-_57222827 | 3.10 |
ENSMUST00000177425.1
|
C3
|
complement component 3 |
| chr6_-_126166726 | 3.07 |
ENSMUST00000112244.2
ENSMUST00000050484.7 |
Ntf3
|
neurotrophin 3 |
| chr16_+_44173239 | 3.07 |
ENSMUST00000119746.1
|
Gm608
|
predicted gene 608 |
| chr2_+_33216051 | 3.07 |
ENSMUST00000004208.5
|
Angptl2
|
angiopoietin-like 2 |
| chr1_+_93235836 | 3.06 |
ENSMUST00000062202.7
|
Sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr7_+_26808880 | 3.05 |
ENSMUST00000040944.7
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr11_+_28853189 | 3.00 |
ENSMUST00000020759.5
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr4_+_135759705 | 2.97 |
ENSMUST00000105854.1
|
Myom3
|
myomesin family, member 3 |
| chr7_-_80403315 | 2.96 |
ENSMUST00000147150.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr7_-_140900401 | 2.95 |
ENSMUST00000026561.8
|
Cox8b
|
cytochrome c oxidase subunit VIIIb |
| chr6_+_138140521 | 2.88 |
ENSMUST00000120939.1
ENSMUST00000120302.1 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr14_+_65666394 | 2.88 |
ENSMUST00000022610.8
|
Scara5
|
scavenger receptor class A, member 5 (putative) |
| chr15_-_78206391 | 2.87 |
ENSMUST00000120592.1
|
Pvalb
|
parvalbumin |
| chr10_+_75406911 | 2.87 |
ENSMUST00000039925.7
|
Upb1
|
ureidopropionase, beta |
| chr8_+_45658666 | 2.86 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_124542281 | 2.84 |
ENSMUST00000159463.1
ENSMUST00000162844.1 ENSMUST00000160505.1 ENSMUST00000162443.1 |
C1s
|
complement component 1, s subcomponent |
| chr8_-_83955205 | 2.80 |
ENSMUST00000098595.2
|
Gm10644
|
predicted gene 10644 |
| chr7_+_27195781 | 2.79 |
ENSMUST00000108379.1
ENSMUST00000179391.1 |
BC024978
|
cDNA sequence BC024978 |
| chr17_-_24209377 | 2.79 |
ENSMUST00000024931.4
|
Ntn3
|
netrin 3 |
| chr11_+_99047311 | 2.77 |
ENSMUST00000140772.1
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr17_-_34959232 | 2.77 |
ENSMUST00000165202.1
ENSMUST00000172753.1 |
Hspa1b
|
heat shock protein 1B |
| chr6_-_54992946 | 2.76 |
ENSMUST00000131475.1
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chr1_+_152399824 | 2.75 |
ENSMUST00000044311.8
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chrX_+_38316177 | 2.75 |
ENSMUST00000016471.2
ENSMUST00000115134.1 |
Atp1b4
|
ATPase, (Na+)/K+ transporting, beta 4 polypeptide |
| chr17_-_6948283 | 2.73 |
ENSMUST00000024572.9
|
Rsph3b
|
radial spoke 3B homolog (Chlamydomonas) |
| chr6_+_113471481 | 2.73 |
ENSMUST00000113062.1
|
Il17rc
|
interleukin 17 receptor C |
| chr4_+_42629719 | 2.72 |
ENSMUST00000166898.2
|
Gm2564
|
predicted gene 2564 |
| chr1_+_127729405 | 2.71 |
ENSMUST00000038006.6
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr10_+_62071014 | 2.70 |
ENSMUST00000053865.5
|
Gm5424
|
predicted gene 5424 |
| chr3_+_107631322 | 2.69 |
ENSMUST00000106703.1
|
Gm10961
|
predicted gene 10961 |
| chr17_-_37023349 | 2.66 |
ENSMUST00000102665.4
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr6_+_124304646 | 2.65 |
ENSMUST00000112541.2
ENSMUST00000032234.2 |
Cd163
|
CD163 antigen |
| chr6_+_113471427 | 2.62 |
ENSMUST00000058300.7
|
Il17rc
|
interleukin 17 receptor C |
| chr1_-_162859684 | 2.62 |
ENSMUST00000131058.1
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr10_-_95415484 | 2.62 |
ENSMUST00000172070.1
ENSMUST00000150432.1 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr17_-_34972124 | 2.61 |
ENSMUST00000087328.2
ENSMUST00000179128.1 |
Hspa1a
|
heat shock protein 1A |
| chrX_-_75875101 | 2.59 |
ENSMUST00000114059.3
|
Pls3
|
plastin 3 (T-isoform) |
| chr5_-_31295862 | 2.58 |
ENSMUST00000041266.7
ENSMUST00000172435.1 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr4_+_102570065 | 2.55 |
ENSMUST00000097950.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_77170534 | 2.55 |
ENSMUST00000111833.2
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr2_-_148045891 | 2.53 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr6_-_59024340 | 2.53 |
ENSMUST00000173193.1
|
Fam13a
|
family with sequence similarity 13, member A |
| chr15_-_34495180 | 2.52 |
ENSMUST00000022946.5
|
Hrsp12
|
heat-responsive protein 12 |
| chr3_+_89229046 | 2.51 |
ENSMUST00000041142.3
|
Muc1
|
mucin 1, transmembrane |
| chr11_-_94601862 | 2.49 |
ENSMUST00000103164.3
|
Acsf2
|
acyl-CoA synthetase family member 2 |
| chr2_+_25054396 | 2.48 |
ENSMUST00000102931.4
ENSMUST00000074422.7 ENSMUST00000132172.1 ENSMUST00000114388.1 ENSMUST00000114386.1 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr8_-_105943382 | 2.47 |
ENSMUST00000038896.7
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr9_+_74953053 | 2.47 |
ENSMUST00000170846.1
|
Fam214a
|
family with sequence similarity 214, member A |
| chr10_-_89506631 | 2.47 |
ENSMUST00000058126.8
ENSMUST00000105296.2 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_-_104876383 | 2.46 |
ENSMUST00000064873.8
ENSMUST00000106808.3 ENSMUST00000048947.8 |
C8a
|
complement component 8, alpha polypeptide |
| chrX_-_48208566 | 2.46 |
ENSMUST00000037960.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr19_+_16435616 | 2.46 |
ENSMUST00000025602.2
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chrX_-_73716145 | 2.46 |
ENSMUST00000002091.5
|
Bcap31
|
B cell receptor associated protein 31 |
| chr9_+_7445822 | 2.44 |
ENSMUST00000034497.6
|
Mmp3
|
matrix metallopeptidase 3 |
| chr7_+_119617781 | 2.44 |
ENSMUST00000047929.6
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr3_-_107943390 | 2.43 |
ENSMUST00000106681.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr8_+_45658273 | 2.43 |
ENSMUST00000153798.1
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr4_-_135494499 | 2.42 |
ENSMUST00000105856.2
|
Nipal3
|
NIPA-like domain containing 3 |
| chr1_-_13660476 | 2.40 |
ENSMUST00000027071.5
|
Lactb2
|
lactamase, beta 2 |
| chr11_-_106579111 | 2.39 |
ENSMUST00000103070.2
|
Tex2
|
testis expressed gene 2 |
| chr7_+_119900099 | 2.39 |
ENSMUST00000106516.1
|
Lyrm1
|
LYR motif containing 1 |
| chr2_+_133552159 | 2.39 |
ENSMUST00000028836.6
|
Bmp2
|
bone morphogenetic protein 2 |
| chr6_+_90333270 | 2.39 |
ENSMUST00000164761.1
ENSMUST00000046128.9 |
Uroc1
|
urocanase domain containing 1 |
| chr17_-_31637135 | 2.39 |
ENSMUST00000118504.1
ENSMUST00000078509.5 ENSMUST00000067801.6 |
Cbs
|
cystathionine beta-synthase |
| chr19_-_29047847 | 2.38 |
ENSMUST00000025696.4
|
Ak3
|
adenylate kinase 3 |
| chr5_-_65435717 | 2.37 |
ENSMUST00000117542.1
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr17_-_73950172 | 2.36 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr11_-_77894096 | 2.35 |
ENSMUST00000017597.4
|
Pipox
|
pipecolic acid oxidase |
| chr4_+_43632185 | 2.35 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr7_-_126897424 | 2.35 |
ENSMUST00000120007.1
|
Tmem219
|
transmembrane protein 219 |
| chr19_-_4698668 | 2.33 |
ENSMUST00000177696.1
|
Gm960
|
predicted gene 960 |
| chr15_-_74728011 | 2.33 |
ENSMUST00000023261.2
|
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr2_+_32646586 | 2.33 |
ENSMUST00000009705.7
ENSMUST00000167841.1 |
Eng
|
endoglin |
| chr10_-_75773350 | 2.33 |
ENSMUST00000001716.7
|
Ddt
|
D-dopachrome tautomerase |
| chr2_+_69135799 | 2.32 |
ENSMUST00000041865.7
|
Nostrin
|
nitric oxide synthase trafficker |
| chr1_+_164796723 | 2.32 |
ENSMUST00000027861.4
|
Dpt
|
dermatopontin |
| chr2_+_55437100 | 2.31 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr6_+_90465287 | 2.30 |
ENSMUST00000113530.1
|
Klf15
|
Kruppel-like factor 15 |
| chr10_-_95415283 | 2.30 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr5_-_65435881 | 2.30 |
ENSMUST00000031103.7
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr14_+_32028989 | 2.29 |
ENSMUST00000022460.4
|
Galnt15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr9_-_103365769 | 2.28 |
ENSMUST00000035484.4
ENSMUST00000072249.6 |
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr16_+_43510267 | 2.27 |
ENSMUST00000114695.2
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_+_20007938 | 2.27 |
ENSMUST00000125799.1
ENSMUST00000121491.1 |
Ttpa
|
tocopherol (alpha) transfer protein |
| chr17_+_80944611 | 2.26 |
ENSMUST00000025092.4
|
Tmem178
|
transmembrane protein 178 |
| chr19_-_6067785 | 2.25 |
ENSMUST00000162575.1
ENSMUST00000159084.1 ENSMUST00000161718.1 ENSMUST00000162810.1 ENSMUST00000025713.5 ENSMUST00000113543.2 ENSMUST00000161528.1 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr10_-_81545175 | 2.25 |
ENSMUST00000043604.5
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr19_-_24861828 | 2.25 |
ENSMUST00000047666.4
|
Pgm5
|
phosphoglucomutase 5 |
| chr2_-_160327494 | 2.25 |
ENSMUST00000099127.2
|
Gm826
|
predicted gene 826 |
| chr12_-_86892540 | 2.24 |
ENSMUST00000181290.1
|
Gm26698
|
predicted gene, 26698 |
| chr13_+_38345716 | 2.23 |
ENSMUST00000171970.1
|
Bmp6
|
bone morphogenetic protein 6 |
| chr19_-_5457397 | 2.23 |
ENSMUST00000179549.1
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr7_+_28179469 | 2.23 |
ENSMUST00000085901.6
ENSMUST00000172761.1 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr6_+_113483297 | 2.23 |
ENSMUST00000032422.5
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr17_+_35439155 | 2.22 |
ENSMUST00000071951.6
ENSMUST00000078205.7 ENSMUST00000116598.3 ENSMUST00000076256.7 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr9_+_107576915 | 2.22 |
ENSMUST00000112387.2
ENSMUST00000123005.1 ENSMUST00000010195.7 ENSMUST00000144392.1 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr4_-_154899077 | 2.21 |
ENSMUST00000030935.3
ENSMUST00000132281.1 |
Fam213b
|
family with sequence similarity 213, member B |
| chr11_+_78290841 | 2.21 |
ENSMUST00000046361.4
|
BC030499
|
cDNA sequence BC030499 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfia
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.6 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 4.0 | 11.9 | GO:1902617 | response to fluoride(GO:1902617) |
| 3.3 | 10.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 3.0 | 9.0 | GO:1903918 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 2.5 | 14.7 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 2.4 | 9.5 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 2.2 | 24.6 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 2.2 | 11.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 2.0 | 11.9 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.9 | 5.6 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.9 | 13.0 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.7 | 3.4 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.7 | 5.1 | GO:0009804 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 1.6 | 6.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.6 | 6.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.5 | 7.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.5 | 4.4 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 1.4 | 4.2 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 1.3 | 4.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.3 | 5.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.2 | 7.4 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.2 | 2.4 | GO:0003133 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 1.1 | 4.5 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 1.1 | 12.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.1 | 3.3 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 1.1 | 3.2 | GO:0006507 | GPI anchor release(GO:0006507) |
| 1.1 | 6.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.0 | 6.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 1.0 | 3.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.0 | 9.0 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.9 | 4.5 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.9 | 4.5 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.9 | 0.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.9 | 3.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 8.7 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.9 | 4.3 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.8 | 1.7 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.8 | 1.7 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.8 | 4.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.8 | 7.8 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.8 | 2.3 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.8 | 4.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.7 | 2.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.7 | 3.0 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.7 | 8.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.7 | 1.5 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.7 | 4.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.7 | 7.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.7 | 2.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 5.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.7 | 5.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.7 | 2.0 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.6 | 6.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.6 | 10.6 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.6 | 1.9 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.6 | 3.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.6 | 1.2 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.6 | 3.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.6 | 1.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.6 | 2.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.6 | 4.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.6 | 1.7 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.6 | 1.7 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.6 | 2.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.6 | 0.6 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.5 | 1.6 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.5 | 2.1 | GO:0006001 | fructose catabolic process(GO:0006001) NADH oxidation(GO:0006116) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.5 | 1.6 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.5 | 1.6 | GO:0061076 | negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.5 | 1.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 4.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.5 | 1.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.5 | 2.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 1.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) ammonia assimilation cycle(GO:0019676) |
| 0.5 | 2.0 | GO:0060397 | positive regulation of skeletal muscle cell proliferation(GO:0014858) JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.5 | 2.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.5 | 1.5 | GO:0046439 | cysteine catabolic process(GO:0009093) glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-cysteine catabolic process(GO:0019448) L-alanine catabolic process(GO:0042853) L-cysteine metabolic process(GO:0046439) |
| 0.5 | 1.5 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.5 | 2.4 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.5 | 1.9 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.5 | 2.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.5 | 1.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 1.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.4 | 1.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.4 | 1.8 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.4 | 1.8 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.4 | 2.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.4 | 0.9 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.4 | 3.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.4 | 0.9 | GO:0072199 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.4 | 14.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.4 | 0.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.4 | 1.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.4 | 1.7 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.4 | 2.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.4 | 1.3 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.4 | 3.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.4 | 4.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.4 | 14.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.4 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.6 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.4 | 4.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.4 | 2.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.4 | 0.8 | GO:0046113 | purine nucleobase catabolic process(GO:0006145) nucleobase catabolic process(GO:0046113) |
| 0.4 | 3.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.4 | 1.5 | GO:0015744 | succinate transport(GO:0015744) |
| 0.4 | 2.7 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 | 1.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.4 | 5.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.4 | 1.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 1.9 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.4 | 3.7 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.4 | 3.4 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.4 | 6.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 1.8 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.4 | 1.1 | GO:0070256 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.4 | 3.6 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.4 | 0.4 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.3 | 5.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.3 | 1.4 | GO:1990401 | embryonic lung development(GO:1990401) |
| 0.3 | 1.4 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.3 | 1.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 2.0 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.3 | 1.0 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.3 | 2.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 1.0 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.3 | 1.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.3 | 1.3 | GO:0009217 | purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.3 | 0.9 | GO:0090265 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.3 | 1.3 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.3 | 0.9 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 0.6 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.3 | 0.9 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 0.3 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.3 | 1.5 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 5.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 0.6 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.3 | 1.7 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.3 | 2.0 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.3 | 0.9 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.3 | 2.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.3 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 3.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 13.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.6 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.3 | 0.8 | GO:0035788 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) regulation of metanephric mesenchymal cell migration(GO:2000589) |
| 0.3 | 1.1 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.3 | 1.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.3 | 1.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 0.8 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.3 | 1.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 1.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 5.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 2.5 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.2 | 9.4 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 5.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 0.7 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 0.5 | GO:2000974 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 0.9 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 0.7 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.2 | 0.9 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.2 | 0.7 | GO:0015881 | creatine transport(GO:0015881) |
| 0.2 | 0.7 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.2 | 1.5 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.2 | 0.9 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 1.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 4.9 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 1.5 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.2 | 2.1 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 0.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 0.6 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 1.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.2 | 1.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.5 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 1.5 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 1.4 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 1.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 1.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 0.6 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.2 | 0.4 | GO:0071655 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.2 | 0.6 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 1.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.3 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.2 | 0.7 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.2 | 1.9 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 1.5 | GO:0000050 | urea cycle(GO:0000050) |
| 0.2 | 1.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 1.8 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 0.7 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.2 | 1.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.2 | 1.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.6 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.2 | 2.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.5 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 1.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 0.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 0.5 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.5 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 1.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 | 1.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.9 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 | 0.3 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.2 | 0.3 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.2 | 0.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 0.9 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.2 | 2.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 0.7 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.2 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.2 | 0.5 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 8.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 3.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.0 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.2 | 0.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 1.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.2 | GO:2000911 | positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.2 | 0.5 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 3.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 0.6 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 0.5 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 3.8 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.2 | 3.3 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.2 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 1.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.7 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.5 | GO:0034033 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 1.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.6 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 1.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.6 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 | 0.3 | GO:1905204 | negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 | 2.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 1.2 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 1.4 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.8 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.8 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.8 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 1.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.6 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 3.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 1.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.7 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 2.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.6 | GO:1903609 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 8.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.1 | 1.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.5 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.1 | 0.2 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 1.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 1.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 2.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.3 | GO:2000757 | negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.1 | 0.4 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 1.8 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 1.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.7 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 2.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.9 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.5 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.1 | 0.8 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 3.7 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 5.4 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 0.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 2.0 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.1 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 5.3 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 2.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.5 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 0.3 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.1 | 0.9 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.8 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.9 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.4 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 1.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.4 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.3 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.7 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.4 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 0.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.8 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.6 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.2 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 0.7 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.5 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 0.7 | GO:0015682 | ferric iron transport(GO:0015682) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.0 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.5 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 2.9 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.5 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.4 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.3 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 7.4 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.1 | 0.9 | GO:2000191 | regulation of fatty acid transport(GO:2000191) |
| 0.1 | 3.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.4 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.8 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.4 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 4.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.5 | GO:0060903 | germ cell migration(GO:0008354) positive regulation of meiosis I(GO:0060903) |
| 0.1 | 1.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 5.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.2 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 2.8 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.1 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.0 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0060717 | chorion development(GO:0060717) |
| 0.1 | 0.9 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.1 | 0.3 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.1 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 1.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.7 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 0.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 0.5 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 1.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 1.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.9 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.9 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 2.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 2.9 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.2 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 1.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.4 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 2.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 2.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.2 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0097647 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 1.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.8 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.4 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.8 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 3.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) regulation of sulfur metabolic process(GO:0042762) positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.7 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 1.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.5 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.3 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 2.1 | 2.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 1.3 | 20.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.3 | 9.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.0 | 11.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 1.9 | GO:0044317 | rod spherule(GO:0044317) |
| 0.6 | 2.5 | GO:0036019 | endolysosome(GO:0036019) |
| 0.6 | 1.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.6 | 6.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 2.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.6 | 2.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 1.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.5 | 2.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.5 | 3.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.5 | 7.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 3.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.4 | 1.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 2.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 3.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 8.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 1.5 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.4 | 1.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.4 | 2.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 2.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 1.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 1.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 2.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.3 | 1.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.3 | 0.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 1.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.3 | 2.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 0.9 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 4.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 2.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 4.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 5.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 0.9 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 0.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 1.6 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 2.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 8.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.2 | 36.3 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.2 | 0.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 1.7 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 6.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 5.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 0.6 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 4.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 3.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 3.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 2.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 4.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 14.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 5.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 0.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 0.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 2.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 2.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 2.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 2.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 1.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 9.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 2.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 3.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 5.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 2.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.4 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 3.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 14.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 12.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 2.4 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 58.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 31.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 3.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 22.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 4.1 | 16.6 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 4.0 | 11.9 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 4.0 | 11.9 | GO:0005186 | pheromone activity(GO:0005186) |
| 3.1 | 9.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 3.1 | 9.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 2.5 | 10.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.1 | 21.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 2.1 | 12.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 2.0 | 6.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.9 | 7.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.9 | 7.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.8 | 5.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.6 | 4.7 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 1.5 | 9.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 1.5 | 4.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 1.3 | 6.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.1 | 3.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.0 | 3.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 1.0 | 10.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.0 | 5.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 1.0 | 5.0 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 1.0 | 4.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.0 | 4.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.0 | 4.8 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.9 | 19.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.9 | 5.6 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.9 | 7.8 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.8 | 4.0 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.8 | 2.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.8 | 3.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.8 | 23.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.8 | 3.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.8 | 2.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.8 | 2.3 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.7 | 3.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.7 | 1.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.7 | 2.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.7 | 4.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.7 | 5.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.7 | 2.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.6 | 2.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.6 | 2.3 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.6 | 5.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.6 | 2.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 5.9 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.5 | 5.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 6.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.5 | 8.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.5 | 1.5 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.5 | 1.5 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.5 | 2.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 3.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 1.5 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.5 | 2.0 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.5 | 3.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.5 | 2.4 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.5 | 2.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 2.7 | GO:0008410 | acetyl-CoA C-acetyltransferase activity(GO:0003985) CoA-transferase activity(GO:0008410) palmitoyl-CoA oxidase activity(GO:0016401) C-acetyltransferase activity(GO:0016453) |
| 0.5 | 17.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 9.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 3.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.4 | 2.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 2.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.4 | 2.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.4 | 1.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.4 | 1.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.4 | 2.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.2 | GO:0017099 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.4 | 1.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.4 | 1.5 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.4 | 11.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 1.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 1.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 5.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 2.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.3 | 3.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 1.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.3 | 2.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 2.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 1.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 1.5 | GO:0070404 | NADH binding(GO:0070404) |
| 0.3 | 7.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 0.9 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.3 | 1.2 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.3 | 1.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 1.5 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.3 | 2.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 3.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 4.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 5.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 0.8 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.3 | 1.3 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 3.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 1.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 1.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 2.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 1.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 1.7 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 2.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 1.7 | GO:0016812 | nucleobase binding(GO:0002054) hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 0.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 1.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 4.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 1.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 0.9 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.2 | 6.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 0.7 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.2 | 0.9 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.2 | 4.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 2.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.6 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 2.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.2 | 3.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 2.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 0.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 0.6 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 7.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.6 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.2 | 2.6 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.2 | 4.4 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.2 | 1.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 7.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 1.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 1.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 1.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 2.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 28.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.2 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 2.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 0.5 | GO:0071820 | N-box binding(GO:0071820) |
| 0.2 | 2.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 3.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 1.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 2.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.9 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.5 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.7 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.4 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 3.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 5.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 5.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 4.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.6 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.5 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 1.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.1 | 0.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 3.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 3.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 1.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.8 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 4.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.4 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 4.8 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 6.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 1.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) primary miRNA binding(GO:0070878) |
| 0.1 | 0.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 2.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 3.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 2.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 9.0 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 7.6 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.7 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 2.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.4 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 32.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 13.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 3.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 9.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 10.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 7.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 6.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 5.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 9.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 5.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 5.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 7.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 4.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 0.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 1.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.5 | 4.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.4 | 15.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.9 | 12.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.8 | 15.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.7 | 12.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 7.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.6 | 28.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.4 | 15.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 4.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 37.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.4 | 6.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 4.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 4.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 3.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 4.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 2.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.3 | 7.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 2.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 6.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 9.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 6.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 2.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 5.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 2.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 11.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 3.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 1.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 2.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.1 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 2.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 3.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 2.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 4.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.4 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.1 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 3.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.6 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.8 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.0 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |