Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
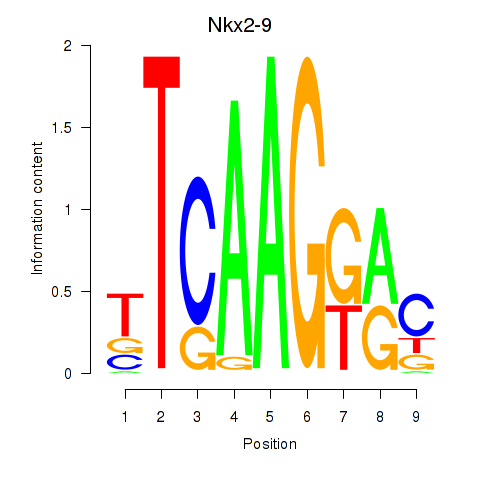
Results for Nkx2-9
Z-value: 0.55
Transcription factors associated with Nkx2-9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-9
|
ENSMUSG00000058669.7 | NK2 homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-9 | mm10_v2_chr12_-_56613270_56613291 | 0.02 | 9.1e-01 | Click! |
Activity profile of Nkx2-9 motif
Sorted Z-values of Nkx2-9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_98490522 | 4.87 |
ENSMUST00000035029.2
|
Rbp2
|
retinol binding protein 2, cellular |
| chr6_-_40999479 | 4.32 |
ENSMUST00000166306.1
|
Gm2663
|
predicted gene 2663 |
| chr6_+_41354105 | 3.90 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr6_+_78370877 | 3.55 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr7_-_142576492 | 3.51 |
ENSMUST00000140716.1
|
H19
|
H19 fetal liver mRNA |
| chr4_-_119189949 | 3.47 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr2_+_173022360 | 3.00 |
ENSMUST00000173997.1
|
Rbm38
|
RNA binding motif protein 38 |
| chr6_-_78378851 | 2.83 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr4_-_119190005 | 2.16 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr6_+_145121727 | 1.92 |
ENSMUST00000032396.6
|
Lrmp
|
lymphoid-restricted membrane protein |
| chrX_+_135993820 | 1.83 |
ENSMUST00000058119.7
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr2_+_103969528 | 1.65 |
ENSMUST00000123437.1
ENSMUST00000163256.1 |
Lmo2
|
LIM domain only 2 |
| chr15_-_79285502 | 1.61 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr4_+_34893772 | 1.61 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr10_+_21690845 | 1.51 |
ENSMUST00000071008.3
|
Gm5420
|
predicted gene 5420 |
| chr7_+_30422389 | 1.44 |
ENSMUST00000108175.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr6_-_78468863 | 1.42 |
ENSMUST00000032089.2
|
Reg3g
|
regenerating islet-derived 3 gamma |
| chr12_-_114416895 | 1.40 |
ENSMUST00000179796.1
|
Ighv6-5
|
immunoglobulin heavy variable V6-5 |
| chr11_+_117849223 | 1.39 |
ENSMUST00000081387.4
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chrX_-_49788204 | 1.37 |
ENSMUST00000114893.1
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr9_+_57504012 | 1.31 |
ENSMUST00000080514.7
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr9_-_75683992 | 1.31 |
ENSMUST00000034699.6
|
Scg3
|
secretogranin III |
| chr10_-_86732409 | 1.31 |
ENSMUST00000070435.4
|
Fabp3-ps1
|
fatty acid binding protein 3, muscle and heart, pseudogene 1 |
| chr3_+_96670131 | 1.26 |
ENSMUST00000048427.5
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr11_+_117849286 | 1.05 |
ENSMUST00000093906.4
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr6_-_113434529 | 1.00 |
ENSMUST00000133348.1
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr4_-_138757578 | 0.91 |
ENSMUST00000030526.6
|
Pla2g2f
|
phospholipase A2, group IIF |
| chr6_+_78380700 | 0.88 |
ENSMUST00000101272.1
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr14_+_77156733 | 0.78 |
ENSMUST00000022589.7
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr3_-_59101810 | 0.78 |
ENSMUST00000085040.4
|
Gpr171
|
G protein-coupled receptor 171 |
| chr6_+_135362931 | 0.77 |
ENSMUST00000032330.9
|
Emp1
|
epithelial membrane protein 1 |
| chr17_+_32403006 | 0.75 |
ENSMUST00000065921.5
|
A530088E08Rik
|
RIKEN cDNA A530088E08 gene |
| chr11_-_99493112 | 0.73 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr1_-_181842334 | 0.72 |
ENSMUST00000005003.6
|
Lbr
|
lamin B receptor |
| chr13_-_51567084 | 0.72 |
ENSMUST00000021898.5
|
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr9_-_58741543 | 0.72 |
ENSMUST00000098674.4
|
2410076I21Rik
|
RIKEN cDNA 2410076I21 gene |
| chr18_-_35662180 | 0.71 |
ENSMUST00000025209.4
ENSMUST00000096573.2 |
Spata24
|
spermatogenesis associated 24 |
| chr6_-_142507805 | 0.68 |
ENSMUST00000134191.1
ENSMUST00000032373.5 |
Ldhb
|
lactate dehydrogenase B |
| chr6_-_136401830 | 0.68 |
ENSMUST00000058713.7
|
E330021D16Rik
|
RIKEN cDNA E330021D16 gene |
| chr10_+_88091070 | 0.66 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr4_+_141420757 | 0.64 |
ENSMUST00000102486.4
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr8_+_94377911 | 0.63 |
ENSMUST00000159142.1
|
Gm15889
|
predicted gene 15889 |
| chr1_-_120505084 | 0.63 |
ENSMUST00000027639.1
|
Marco
|
macrophage receptor with collagenous structure |
| chr1_+_63176818 | 0.59 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chrX_+_56779437 | 0.55 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chrX_+_13632769 | 0.55 |
ENSMUST00000096492.3
|
Gpr34
|
G protein-coupled receptor 34 |
| chr2_-_152398046 | 0.49 |
ENSMUST00000063332.8
ENSMUST00000182625.1 |
Sox12
|
SRY-box containing gene 12 |
| chr18_-_65393844 | 0.48 |
ENSMUST00000035548.8
|
Alpk2
|
alpha-kinase 2 |
| chr15_-_98777261 | 0.48 |
ENSMUST00000166022.1
|
Wnt10b
|
wingless related MMTV integration site 10b |
| chr10_+_68723723 | 0.47 |
ENSMUST00000080995.6
|
Tmem26
|
transmembrane protein 26 |
| chr1_+_74153981 | 0.46 |
ENSMUST00000027372.7
ENSMUST00000106899.2 |
Cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr10_+_14523062 | 0.46 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr17_-_51833278 | 0.46 |
ENSMUST00000133574.1
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr9_-_56161051 | 0.45 |
ENSMUST00000034876.8
|
Tspan3
|
tetraspanin 3 |
| chr12_-_24493656 | 0.45 |
ENSMUST00000073088.2
|
Gm16372
|
predicted pseudogene 16372 |
| chrX_-_102189371 | 0.45 |
ENSMUST00000033683.7
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr8_+_95703037 | 0.44 |
ENSMUST00000073139.7
ENSMUST00000080666.7 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr15_+_79690869 | 0.43 |
ENSMUST00000046463.8
|
Gtpbp1
|
GTP binding protein 1 |
| chr8_+_95633500 | 0.40 |
ENSMUST00000034094.9
|
Gins3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr12_+_69790288 | 0.39 |
ENSMUST00000021378.3
|
4930512B01Rik
|
RIKEN cDNA 4930512B01 gene |
| chr7_+_30280094 | 0.36 |
ENSMUST00000108187.1
ENSMUST00000014072.5 |
Thap8
|
THAP domain containing 8 |
| chr15_-_78204228 | 0.36 |
ENSMUST00000005860.9
|
Pvalb
|
parvalbumin |
| chrX_-_6320717 | 0.35 |
ENSMUST00000024049.7
|
Bmp15
|
bone morphogenetic protein 15 |
| chr3_+_37348645 | 0.35 |
ENSMUST00000038885.3
|
Fgf2
|
fibroblast growth factor 2 |
| chr2_+_80638798 | 0.35 |
ENSMUST00000028382.6
ENSMUST00000124377.1 |
Nup35
|
nucleoporin 35 |
| chr5_+_147430407 | 0.33 |
ENSMUST00000176600.1
|
Pan3
|
PAN3 polyA specific ribonuclease subunit homolog (S. cerevisiae) |
| chr11_-_106788845 | 0.33 |
ENSMUST00000123339.1
|
Ddx5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 |
| chr6_+_35177610 | 0.32 |
ENSMUST00000170234.1
|
Nup205
|
nucleoporin 205 |
| chr14_+_75455957 | 0.31 |
ENSMUST00000164848.1
|
Siah3
|
seven in absentia homolog 3 (Drosophila) |
| chr4_+_155891822 | 0.31 |
ENSMUST00000105584.3
ENSMUST00000079031.5 |
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr11_-_106789157 | 0.30 |
ENSMUST00000129585.1
|
Ddx5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 |
| chr11_+_114668524 | 0.30 |
ENSMUST00000106602.3
ENSMUST00000077915.3 ENSMUST00000106599.1 ENSMUST00000082092.4 |
Rpl38
|
ribosomal protein L38 |
| chr3_-_89245159 | 0.29 |
ENSMUST00000090924.6
|
Trim46
|
tripartite motif-containing 46 |
| chr4_-_41275091 | 0.29 |
ENSMUST00000030143.6
ENSMUST00000108068.1 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr7_+_4925802 | 0.27 |
ENSMUST00000057612.7
|
Ssc5d
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chrX_-_95658392 | 0.25 |
ENSMUST00000120620.1
|
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr6_+_35177386 | 0.25 |
ENSMUST00000043815.9
|
Nup205
|
nucleoporin 205 |
| chr9_-_107770945 | 0.24 |
ENSMUST00000183248.1
ENSMUST00000182022.1 ENSMUST00000035199.6 ENSMUST00000182659.1 |
Rbm5
|
RNA binding motif protein 5 |
| chr16_-_33380717 | 0.23 |
ENSMUST00000180923.1
|
1700007L15Rik
|
RIKEN cDNA 1700007L15 gene |
| chr1_-_135688094 | 0.23 |
ENSMUST00000112103.1
|
Nav1
|
neuron navigator 1 |
| chr7_-_104390586 | 0.22 |
ENSMUST00000106828.1
|
Trim30c
|
tripartite motif-containing 30C |
| chr4_-_72852622 | 0.22 |
ENSMUST00000179234.1
ENSMUST00000078617.4 |
Aldoart1
|
aldolase 1 A, retrogene 1 |
| chr16_+_36694024 | 0.22 |
ENSMUST00000119464.1
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr1_-_128103016 | 0.22 |
ENSMUST00000097597.2
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr16_+_36693972 | 0.21 |
ENSMUST00000023617.6
ENSMUST00000089618.3 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr3_+_37420273 | 0.21 |
ENSMUST00000029277.8
|
Spata5
|
spermatogenesis associated 5 |
| chr11_-_106788486 | 0.20 |
ENSMUST00000021062.5
|
Ddx5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 |
| chr18_-_60848911 | 0.20 |
ENSMUST00000177172.1
ENSMUST00000175934.1 ENSMUST00000176630.1 |
Tcof1
|
Treacher Collins Franceschetti syndrome 1, homolog |
| chr4_-_132884509 | 0.20 |
ENSMUST00000030698.4
|
Stx12
|
syntaxin 12 |
| chr5_+_141856692 | 0.19 |
ENSMUST00000074546.6
|
Sdk1
|
sidekick homolog 1 (chicken) |
| chr1_+_9545397 | 0.19 |
ENSMUST00000072079.7
|
Rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr13_+_109260481 | 0.19 |
ENSMUST00000153234.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr8_+_95320440 | 0.19 |
ENSMUST00000162294.1
|
Tepp
|
testis, prostate and placenta expressed |
| chr12_+_71136848 | 0.18 |
ENSMUST00000149564.1
ENSMUST00000045907.8 |
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chrX_-_95658379 | 0.18 |
ENSMUST00000119640.1
|
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr8_-_80880479 | 0.17 |
ENSMUST00000034150.8
|
Gab1
|
growth factor receptor bound protein 2-associated protein 1 |
| chr4_-_141664063 | 0.16 |
ENSMUST00000084203.4
|
Plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr6_-_34977999 | 0.16 |
ENSMUST00000044387.7
|
2010107G12Rik
|
RIKEN cDNA 2010107G12 gene |
| chr6_-_6217023 | 0.16 |
ENSMUST00000015256.8
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr17_-_25861456 | 0.15 |
ENSMUST00000079461.8
ENSMUST00000176923.1 |
Wdr90
|
WD repeat domain 90 |
| chr2_-_26910569 | 0.14 |
ENSMUST00000015920.5
ENSMUST00000139815.1 ENSMUST00000102899.3 |
Med22
|
mediator complex subunit 22 |
| chr13_-_71963713 | 0.14 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chrX_+_42149288 | 0.13 |
ENSMUST00000115073.2
ENSMUST00000115072.1 |
Stag2
|
stromal antigen 2 |
| chr7_-_102018139 | 0.13 |
ENSMUST00000094134.3
|
Il18bp
|
interleukin 18 binding protein |
| chr2_-_33130565 | 0.12 |
ENSMUST00000124000.1
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr11_+_106789235 | 0.12 |
ENSMUST00000103068.3
ENSMUST00000018516.4 |
Cep95
|
centrosomal protein 95 |
| chr1_-_74600564 | 0.12 |
ENSMUST00000127938.1
ENSMUST00000154874.1 |
Rnf25
|
ring finger protein 25 |
| chr3_-_89245005 | 0.12 |
ENSMUST00000107464.1
|
Trim46
|
tripartite motif-containing 46 |
| chr15_+_100469034 | 0.10 |
ENSMUST00000037001.8
|
Letmd1
|
LETM1 domain containing 1 |
| chr9_-_106887000 | 0.10 |
ENSMUST00000055843.7
|
Rbm15b
|
RNA binding motif protein 15B |
| chr11_+_70029742 | 0.09 |
ENSMUST00000132597.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chrX_-_95658416 | 0.08 |
ENSMUST00000044382.6
|
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr1_+_128103297 | 0.08 |
ENSMUST00000036288.4
|
R3hdm1
|
R3H domain containing 1 |
| chr1_-_23102243 | 0.07 |
ENSMUST00000073179.5
|
4933415F23Rik
|
RIKEN cDNA 4933415F23 gene |
| chr11_-_109363654 | 0.07 |
ENSMUST00000070956.3
|
Gm11696
|
predicted gene 11696 |
| chr11_-_96747405 | 0.06 |
ENSMUST00000180492.1
|
2010300F17Rik
|
RIKEN cDNA 2010300F17 gene |
| chr10_-_120979327 | 0.06 |
ENSMUST00000119944.1
ENSMUST00000119093.1 |
Lemd3
|
LEM domain containing 3 |
| chr13_+_72632597 | 0.05 |
ENSMUST00000172353.1
|
Irx2
|
Iroquois related homeobox 2 (Drosophila) |
| chrX_+_42149534 | 0.05 |
ENSMUST00000127618.1
|
Stag2
|
stromal antigen 2 |
| chr11_+_83409137 | 0.05 |
ENSMUST00000021022.3
|
Rasl10b
|
RAS-like, family 10, member B |
| chr3_-_95411176 | 0.05 |
ENSMUST00000177599.1
|
Gm5070
|
predicted gene 5070 |
| chr18_-_67449083 | 0.04 |
ENSMUST00000025408.8
|
Afg3l2
|
AFG3(ATPase family gene 3)-like 2 (yeast) |
| chr5_+_117413977 | 0.04 |
ENSMUST00000180430.1
|
Ksr2
|
kinase suppressor of ras 2 |
| chr4_+_44981389 | 0.04 |
ENSMUST00000045078.6
ENSMUST00000128973.1 ENSMUST00000151148.1 |
Grhpr
|
glyoxylate reductase/hydroxypyruvate reductase |
| chr18_+_77185815 | 0.04 |
ENSMUST00000079618.4
|
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr7_+_44848991 | 0.03 |
ENSMUST00000107885.1
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr5_+_30105161 | 0.03 |
ENSMUST00000058045.4
|
Gareml
|
GRB2 associated, regulator of MAPK1-like |
| chr13_-_64497792 | 0.03 |
ENSMUST00000180282.1
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene |
| chr11_-_69549108 | 0.03 |
ENSMUST00000108659.1
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr5_-_62765618 | 0.02 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr17_-_29519790 | 0.02 |
ENSMUST00000168877.1
|
Gm17657
|
predicted gene, 17657 |
| chr19_-_43752924 | 0.01 |
ENSMUST00000045562.5
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr1_+_87264345 | 0.01 |
ENSMUST00000118687.1
ENSMUST00000027472.6 |
Efhd1
|
EF hand domain containing 1 |
| chr16_+_30065333 | 0.00 |
ENSMUST00000023171.7
|
Hes1
|
hairy and enhancer of split 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 2.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.4 | 3.5 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.3 | 3.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 0.5 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.2 | 3.0 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 0.7 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.2 | 2.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 1.4 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.5 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 1.6 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 1.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.3 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 | 0.7 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 0.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 1.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.2 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.0 | 0.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.9 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 3.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.4 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.2 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.2 | 1.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 3.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.4 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 4.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 3.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.3 | 0.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 0.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 1.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 8.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 3.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 3.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 4.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 4.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |