Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
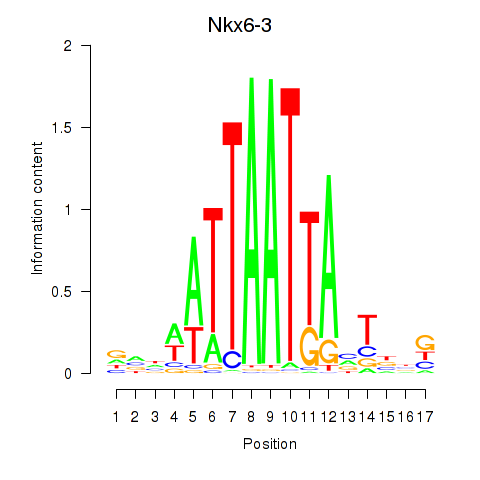
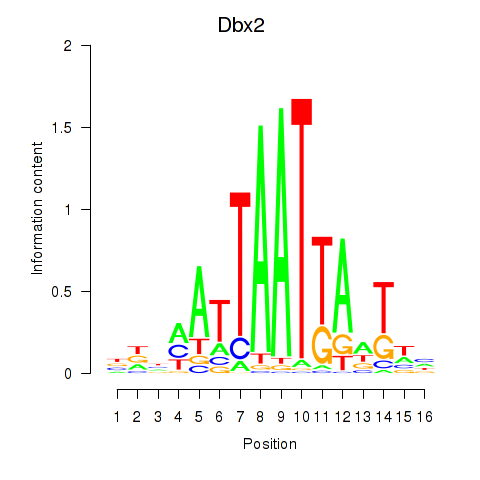
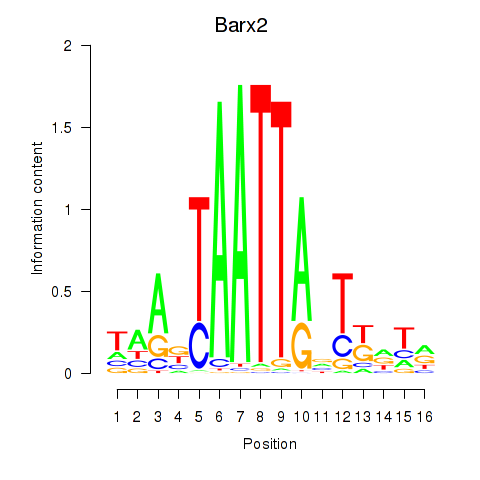
Results for Nkx6-3_Dbx2_Barx2
Z-value: 0.70
Transcription factors associated with Nkx6-3_Dbx2_Barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-3
|
ENSMUSG00000063672.6 | NK6 homeobox 3 |
|
Dbx2
|
ENSMUSG00000045608.6 | developing brain homeobox 2 |
|
Barx2
|
ENSMUSG00000032033.10 | BarH-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dbx2 | mm10_v2_chr15_-_95655960_95655960 | 0.21 | 2.2e-01 | Click! |
| Barx2 | mm10_v2_chr9_-_31913462_31913483 | 0.10 | 5.7e-01 | Click! |
| Nkx6-3 | mm10_v2_chr8_+_23153271_23153271 | 0.06 | 7.2e-01 | Click! |
Activity profile of Nkx6-3_Dbx2_Barx2 motif
Sorted Z-values of Nkx6-3_Dbx2_Barx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_27000362 | 6.77 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr14_+_80000292 | 4.30 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr1_+_174172738 | 3.65 |
ENSMUST00000027817.7
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr1_+_139454747 | 2.69 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr2_-_160619971 | 2.47 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr7_+_43427622 | 2.44 |
ENSMUST00000177164.2
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr17_-_48432723 | 2.42 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr2_-_168767136 | 2.40 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr4_+_34893772 | 2.07 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr10_+_37139558 | 1.95 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr7_+_45621805 | 1.94 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr10_-_76110956 | 1.90 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr2_-_168767029 | 1.89 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr5_-_118244861 | 1.61 |
ENSMUST00000117177.1
ENSMUST00000133372.1 ENSMUST00000154786.1 ENSMUST00000121369.1 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr1_-_172027269 | 1.59 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr11_-_11970540 | 1.57 |
ENSMUST00000109653.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr1_+_40439767 | 1.53 |
ENSMUST00000173514.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_+_40439627 | 1.50 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr2_-_121235689 | 1.46 |
ENSMUST00000142400.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr11_-_17008647 | 1.38 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chr2_+_36230426 | 1.35 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr8_-_62123106 | 1.35 |
ENSMUST00000034052.6
ENSMUST00000034054.7 |
Anxa10
|
annexin A10 |
| chr9_+_72806874 | 1.34 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr16_-_76022266 | 1.32 |
ENSMUST00000114240.1
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr10_-_37138863 | 1.30 |
ENSMUST00000092584.5
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr6_+_38109320 | 1.29 |
ENSMUST00000031851.3
|
Tmem213
|
transmembrane protein 213 |
| chr13_-_58354862 | 1.25 |
ENSMUST00000043605.5
|
Kif27
|
kinesin family member 27 |
| chr8_+_94172618 | 1.25 |
ENSMUST00000034214.6
|
Mt2
|
metallothionein 2 |
| chr7_-_133702515 | 1.23 |
ENSMUST00000153698.1
|
Uros
|
uroporphyrinogen III synthase |
| chr2_-_32387760 | 1.20 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr14_+_58893465 | 1.15 |
ENSMUST00000079960.1
|
Rpl13-ps3
|
ribosomal protein L13, pseudogene 3 |
| chr11_-_99244058 | 1.14 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr7_+_30493622 | 1.13 |
ENSMUST00000058280.6
ENSMUST00000133318.1 ENSMUST00000142575.1 ENSMUST00000131040.1 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr6_-_136941887 | 1.02 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr6_-_128275577 | 1.01 |
ENSMUST00000130454.1
|
Tead4
|
TEA domain family member 4 |
| chr2_+_127854628 | 0.98 |
ENSMUST00000028859.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr5_-_43981757 | 0.94 |
ENSMUST00000061299.7
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr17_-_26099257 | 0.92 |
ENSMUST00000053575.3
|
Gm8186
|
predicted gene 8186 |
| chr9_+_119063429 | 0.91 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr3_-_116253467 | 0.90 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr3_-_14778452 | 0.87 |
ENSMUST00000094365.4
|
Car1
|
carbonic anhydrase 1 |
| chr16_+_45224315 | 0.87 |
ENSMUST00000102802.3
ENSMUST00000063654.4 |
Btla
|
B and T lymphocyte associated |
| chr10_+_116143881 | 0.86 |
ENSMUST00000105271.2
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr17_-_50094277 | 0.85 |
ENSMUST00000113195.1
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr18_+_50051702 | 0.85 |
ENSMUST00000134348.1
ENSMUST00000153873.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_-_136941494 | 0.84 |
ENSMUST00000111892.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr15_-_36879816 | 0.83 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr8_-_106573461 | 0.82 |
ENSMUST00000073722.5
|
Gm10073
|
predicted pseudogene 10073 |
| chr6_-_136941694 | 0.78 |
ENSMUST00000032344.5
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr4_+_145585166 | 0.78 |
ENSMUST00000105739.1
ENSMUST00000119718.1 |
Gm13212
|
predicted gene 13212 |
| chr12_+_102128718 | 0.74 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr1_-_4880669 | 0.73 |
ENSMUST00000078030.3
|
Gm6104
|
predicted gene 6104 |
| chr14_+_61309753 | 0.72 |
ENSMUST00000055159.7
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr4_+_13743424 | 0.72 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_-_17230976 | 0.68 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr11_+_31832660 | 0.67 |
ENSMUST00000132857.1
|
Gm12107
|
predicted gene 12107 |
| chr18_-_23981555 | 0.65 |
ENSMUST00000115829.1
|
Zscan30
|
zinc finger and SCAN domain containing 30 |
| chr18_+_11633276 | 0.63 |
ENSMUST00000115861.2
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr19_-_47692042 | 0.61 |
ENSMUST00000026045.7
ENSMUST00000086923.5 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr17_-_81649607 | 0.59 |
ENSMUST00000163680.2
ENSMUST00000086538.3 ENSMUST00000163123.1 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr2_+_110597298 | 0.58 |
ENSMUST00000045972.6
ENSMUST00000111026.2 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr4_+_32623985 | 0.58 |
ENSMUST00000108178.1
|
Casp8ap2
|
caspase 8 associated protein 2 |
| chr6_-_116716888 | 0.57 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr10_-_127030813 | 0.57 |
ENSMUST00000040560.4
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr18_-_24603464 | 0.57 |
ENSMUST00000154205.1
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr17_-_56036546 | 0.56 |
ENSMUST00000003268.9
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr10_+_79996479 | 0.55 |
ENSMUST00000132517.1
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr6_+_30610984 | 0.55 |
ENSMUST00000062758.4
|
Cpa5
|
carboxypeptidase A5 |
| chr13_-_105271039 | 0.53 |
ENSMUST00000069686.6
|
Rnf180
|
ring finger protein 180 |
| chr10_-_127030789 | 0.53 |
ENSMUST00000120547.1
ENSMUST00000152054.1 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr8_-_24725762 | 0.52 |
ENSMUST00000171438.1
ENSMUST00000171611.1 ENSMUST00000033958.7 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr19_+_5474681 | 0.52 |
ENSMUST00000165485.1
ENSMUST00000166253.1 ENSMUST00000167371.1 ENSMUST00000167855.1 ENSMUST00000070118.7 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr16_-_29544852 | 0.52 |
ENSMUST00000039090.8
|
Atp13a4
|
ATPase type 13A4 |
| chr9_+_38877126 | 0.50 |
ENSMUST00000078289.2
|
Olfr926
|
olfactory receptor 926 |
| chr12_-_55014329 | 0.49 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr8_+_57332111 | 0.49 |
ENSMUST00000181638.1
|
5033428I22Rik
|
RIKEN cDNA 5033428I22 gene |
| chr18_+_53551594 | 0.49 |
ENSMUST00000115398.1
|
Prdm6
|
PR domain containing 6 |
| chr18_-_24603791 | 0.46 |
ENSMUST00000070726.3
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr15_-_75905349 | 0.45 |
ENSMUST00000127550.1
|
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrY_+_90785442 | 0.44 |
ENSMUST00000177591.1
ENSMUST00000177671.1 ENSMUST00000179077.1 |
Erdr1
|
erythroid differentiation regulator 1 |
| chr13_+_120308146 | 0.43 |
ENSMUST00000081558.7
|
BC147527
|
cDNA sequence BC147527 |
| chrX_+_9885622 | 0.43 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr4_-_149126688 | 0.42 |
ENSMUST00000030815.2
|
Cort
|
cortistatin |
| chr2_+_163658370 | 0.41 |
ENSMUST00000164399.1
ENSMUST00000064703.6 ENSMUST00000099105.2 ENSMUST00000152418.1 ENSMUST00000126182.1 ENSMUST00000131228.1 |
Pkig
|
protein kinase inhibitor, gamma |
| chr5_-_45169564 | 0.41 |
ENSMUST00000030975.5
|
4930435H24Rik
|
RIKEN cDNA 4930435H24 gene |
| chr1_-_138175126 | 0.40 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr3_+_24333046 | 0.40 |
ENSMUST00000077389.6
|
Gm7536
|
predicted gene 7536 |
| chr19_+_23723279 | 0.39 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr10_+_116966274 | 0.38 |
ENSMUST00000033651.3
|
D630029K05Rik
|
RIKEN cDNA D630029K05 gene |
| chr2_-_140671400 | 0.38 |
ENSMUST00000056760.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr6_-_119963733 | 0.38 |
ENSMUST00000161512.2
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr19_-_55241236 | 0.38 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr17_+_32468462 | 0.37 |
ENSMUST00000003413.6
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr1_-_138175283 | 0.37 |
ENSMUST00000182755.1
ENSMUST00000183262.1 ENSMUST00000027645.7 ENSMUST00000112036.2 ENSMUST00000182283.1 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr1_+_72284367 | 0.36 |
ENSMUST00000027380.5
ENSMUST00000141783.1 |
Tmem169
|
transmembrane protein 169 |
| chr13_+_109926832 | 0.35 |
ENSMUST00000117420.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr6_+_30611029 | 0.35 |
ENSMUST00000115138.1
|
Cpa5
|
carboxypeptidase A5 |
| chr4_-_14621805 | 0.35 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_+_6248659 | 0.35 |
ENSMUST00000181633.1
ENSMUST00000176283.1 ENSMUST00000175814.1 ENSMUST00000181192.1 |
Gm20619
|
predicted gene 20619 |
| chr14_-_75754475 | 0.34 |
ENSMUST00000049168.7
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr13_-_105054895 | 0.32 |
ENSMUST00000063551.5
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein |
| chr18_+_59062462 | 0.31 |
ENSMUST00000058633.2
ENSMUST00000175897.1 ENSMUST00000118510.1 ENSMUST00000175830.1 |
A730017C20Rik
|
RIKEN cDNA A730017C20 gene |
| chr4_-_147809788 | 0.31 |
ENSMUST00000105734.3
ENSMUST00000176201.1 |
Gm13157
Gm20707
|
predicted gene 13157 predicted gene 20707 |
| chr19_-_38043559 | 0.30 |
ENSMUST00000041475.8
ENSMUST00000172095.2 |
Myof
|
myoferlin |
| chr16_+_92478743 | 0.30 |
ENSMUST00000160494.1
|
2410124H12Rik
|
RIKEN cDNA 2410124H12 gene |
| chr11_-_82908360 | 0.30 |
ENSMUST00000103213.3
|
Nle1
|
notchless homolog 1 (Drosophila) |
| chr8_+_34054622 | 0.29 |
ENSMUST00000149618.1
|
Gm9951
|
predicted gene 9951 |
| chr10_+_23797052 | 0.29 |
ENSMUST00000133289.1
|
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr1_-_138175238 | 0.28 |
ENSMUST00000182536.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr7_+_79273201 | 0.28 |
ENSMUST00000037315.6
|
Abhd2
|
abhydrolase domain containing 2 |
| chr4_+_147132038 | 0.28 |
ENSMUST00000084149.3
|
Gm13139
|
predicted gene 13139 |
| chr18_-_43477764 | 0.28 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr4_+_66827545 | 0.27 |
ENSMUST00000048096.5
ENSMUST00000107365.2 |
Tlr4
|
toll-like receptor 4 |
| chr19_+_34100943 | 0.26 |
ENSMUST00000025685.6
|
Lipm
|
lipase, family member M |
| chr7_+_103928825 | 0.26 |
ENSMUST00000106863.1
|
Olfr631
|
olfactory receptor 631 |
| chr8_+_121116163 | 0.26 |
ENSMUST00000054691.6
|
Foxc2
|
forkhead box C2 |
| chr15_+_16778101 | 0.26 |
ENSMUST00000026432.6
|
Cdh9
|
cadherin 9 |
| chrX_+_112093496 | 0.25 |
ENSMUST00000130247.2
ENSMUST00000038546.6 |
Tex16
|
testis expressed gene 16 |
| chr3_+_59925214 | 0.25 |
ENSMUST00000049476.2
|
C130079G13Rik
|
RIKEN cDNA C130079G13 gene |
| chr9_+_53771499 | 0.25 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr2_-_85675173 | 0.25 |
ENSMUST00000099917.1
|
Olfr1006
|
olfactory receptor 1006 |
| chr4_-_129378116 | 0.25 |
ENSMUST00000030610.2
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr2_-_113829069 | 0.25 |
ENSMUST00000024005.7
|
Scg5
|
secretogranin V |
| chr6_-_141773810 | 0.24 |
ENSMUST00000148411.1
|
Gm5724
|
predicted gene 5724 |
| chr19_-_39886730 | 0.22 |
ENSMUST00000168838.1
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr18_+_59062282 | 0.22 |
ENSMUST00000165666.2
|
A730017C20Rik
|
RIKEN cDNA A730017C20 gene |
| chr6_-_122340525 | 0.21 |
ENSMUST00000112600.2
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr6_-_3399545 | 0.21 |
ENSMUST00000120087.3
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr2_-_5895319 | 0.20 |
ENSMUST00000026926.4
ENSMUST00000102981.3 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr15_-_99287174 | 0.20 |
ENSMUST00000109100.1
|
Fam186b
|
family with sequence similarity 186, member B |
| chr15_+_81744848 | 0.19 |
ENSMUST00000109554.1
|
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr15_+_82252397 | 0.19 |
ENSMUST00000136948.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr3_-_88410295 | 0.19 |
ENSMUST00000056370.7
|
Pmf1
|
polyamine-modulated factor 1 |
| chr19_-_32196393 | 0.18 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr13_-_21468474 | 0.17 |
ENSMUST00000068235.4
|
Nkapl
|
NFKB activating protein-like |
| chr8_+_92855319 | 0.17 |
ENSMUST00000046290.1
|
Lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr3_-_72967854 | 0.16 |
ENSMUST00000167334.1
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr8_+_11713259 | 0.16 |
ENSMUST00000134409.1
|
1700128E19Rik
|
RIKEN cDNA 1700128E19 gene |
| chr5_+_123907175 | 0.16 |
ENSMUST00000023869.8
|
Denr
|
density-regulated protein |
| chr10_-_130394241 | 0.15 |
ENSMUST00000094502.4
|
Vmn2r84
|
vomeronasal 2, receptor 84 |
| chr1_-_34439672 | 0.15 |
ENSMUST00000042493.8
|
Ccdc115
|
coiled-coil domain containing 115 |
| chr18_+_55057557 | 0.14 |
ENSMUST00000181765.1
|
Gm4221
|
predicted gene 4221 |
| chr1_+_24177610 | 0.14 |
ENSMUST00000054588.8
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr8_-_66486494 | 0.14 |
ENSMUST00000026681.5
|
Tma16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chrX_+_101429555 | 0.14 |
ENSMUST00000033673.6
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr7_-_100583072 | 0.13 |
ENSMUST00000152876.1
ENSMUST00000150042.1 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr5_+_138187485 | 0.13 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr6_-_137169710 | 0.13 |
ENSMUST00000117919.1
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr10_-_110000219 | 0.12 |
ENSMUST00000032719.7
|
Nav3
|
neuron navigator 3 |
| chrX_-_157415286 | 0.12 |
ENSMUST00000079945.4
ENSMUST00000138396.1 |
Phex
|
phosphate regulating gene with homologies to endopeptidases on the X chromosome (hypophosphatemia, vitamin D resistant rickets) |
| chr12_-_118966395 | 0.12 |
ENSMUST00000035515.4
|
Abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr4_-_146909590 | 0.11 |
ENSMUST00000049821.3
|
Gm21411
|
predicted gene, 21411 |
| chr4_+_146161909 | 0.11 |
ENSMUST00000131932.1
|
Zfp600
|
zinc finger protein 600 |
| chr2_-_45112890 | 0.11 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_169685191 | 0.10 |
ENSMUST00000112104.1
ENSMUST00000112107.1 |
Mid1
|
midline 1 |
| chr8_-_105568298 | 0.10 |
ENSMUST00000005849.5
|
Agrp
|
agouti related protein |
| chr1_+_133309778 | 0.10 |
ENSMUST00000094557.4
ENSMUST00000183457.1 ENSMUST00000183738.1 ENSMUST00000185157.1 ENSMUST00000184603.1 |
Golt1a
Kiss1
GOLT1A
|
golgi transport 1 homolog A (S. cerevisiae) KiSS-1 metastasis-suppressor KISS1 isoform e |
| chr2_+_110721340 | 0.09 |
ENSMUST00000111016.2
|
Muc15
|
mucin 15 |
| chr2_-_80128834 | 0.09 |
ENSMUST00000102654.4
ENSMUST00000102655.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_107337556 | 0.09 |
ENSMUST00000040380.6
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr8_+_107031218 | 0.08 |
ENSMUST00000034388.9
|
Vps4a
|
vacuolar protein sorting 4a (yeast) |
| chr2_+_151494182 | 0.08 |
ENSMUST00000089140.6
|
Nsfl1c
|
NSFL1 (p97) cofactor (p47) |
| chr15_-_8710409 | 0.08 |
ENSMUST00000157065.1
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_-_150255591 | 0.08 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
| chr4_+_146610961 | 0.08 |
ENSMUST00000130825.1
|
Gm13248
|
predicted gene 13248 |
| chr7_+_123123870 | 0.07 |
ENSMUST00000094053.5
|
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr12_-_20900867 | 0.07 |
ENSMUST00000079237.5
|
Zfp125
|
zinc finger protein 125 |
| chr10_-_87008015 | 0.07 |
ENSMUST00000035288.8
|
Stab2
|
stabilin 2 |
| chrX_-_8252304 | 0.07 |
ENSMUST00000115594.1
|
Ftsj1
|
FtsJ homolog 1 (E. coli) |
| chr12_+_72536342 | 0.07 |
ENSMUST00000044352.6
|
Pcnxl4
|
pecanex-like 4 (Drosophila) |
| chr2_-_73580288 | 0.07 |
ENSMUST00000028515.3
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr1_-_127840290 | 0.06 |
ENSMUST00000061512.2
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr1_-_187215454 | 0.06 |
ENSMUST00000183819.1
|
Spata17
|
spermatogenesis associated 17 |
| chr4_+_147305674 | 0.06 |
ENSMUST00000148762.3
|
Gm13151
|
predicted gene 13151 |
| chrX_+_170009892 | 0.06 |
ENSMUST00000180251.1
|
Gm21887
|
predicted gene, 21887 |
| chr18_+_23753708 | 0.06 |
ENSMUST00000115830.1
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_-_23273893 | 0.06 |
ENSMUST00000087556.5
|
Smc5
|
structural maintenance of chromosomes 5 |
| chr9_+_113812547 | 0.05 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr7_-_38019505 | 0.05 |
ENSMUST00000085513.4
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr7_+_19119853 | 0.05 |
ENSMUST00000053109.3
|
Fbxo46
|
F-box protein 46 |
| chr10_+_116018213 | 0.04 |
ENSMUST00000063470.4
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_163085643 | 0.04 |
ENSMUST00000096608.4
|
Mroh9
|
maestro heat-like repeat family member 9 |
| chr15_-_43477036 | 0.04 |
ENSMUST00000100683.2
|
Gm10373
|
predicted gene 10373 |
| chr4_-_134245579 | 0.03 |
ENSMUST00000030644.7
|
Zfp593
|
zinc finger protein 593 |
| chr6_+_24795824 | 0.03 |
ENSMUST00000031693.2
|
Spam1
|
sperm adhesion molecule 1 |
| chr1_-_150164943 | 0.03 |
ENSMUST00000181308.1
|
Gm26687
|
predicted gene, 26687 |
| chr1_-_187215421 | 0.03 |
ENSMUST00000110945.3
ENSMUST00000183931.1 ENSMUST00000027908.6 |
Spata17
|
spermatogenesis associated 17 |
| chr9_-_118014160 | 0.02 |
ENSMUST00000111769.3
|
Zcwpw2
|
zinc finger, CW type with PWWP domain 2 |
| chr4_+_30664587 | 0.01 |
ENSMUST00000128925.1
|
4930556G01Rik
|
RIKEN cDNA 4930556G01 gene |
| chr1_+_34439851 | 0.01 |
ENSMUST00000027303.7
|
Imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr1_+_190928492 | 0.01 |
ENSMUST00000066632.7
ENSMUST00000110899.1 |
Angel2
|
angel homolog 2 (Drosophila) |
| chr12_-_11208948 | 0.01 |
ENSMUST00000049877.1
|
Msgn1
|
mesogenin 1 |
| chr5_-_34660068 | 0.00 |
ENSMUST00000041364.9
|
Nop14
|
NOP14 nucleolar protein |
| chr18_+_24603952 | 0.00 |
ENSMUST00000025120.6
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-3_Dbx2_Barx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.8 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 2.7 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.5 | 1.6 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.4 | 1.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 2.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 1.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.3 | 1.4 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 0.3 | 1.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 3.0 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.2 | 2.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 4.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.2 | 1.0 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 0.6 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.2 | 1.3 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 2.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 0.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 2.1 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 1.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.1 | 0.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.6 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.1 | 1.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.1 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 3.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.3 | GO:0045362 | interleukin-13 biosynthetic process(GO:0042231) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.3 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.6 | GO:0098735 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.6 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 0.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 4.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 1.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.3 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 1.3 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.0 | 0.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.5 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.4 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 2.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.2 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.4 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 2.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 1.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.4 | 1.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 4.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 4.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 2.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 2.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 0.6 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 2.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 6.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 4.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 4.2 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |