Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
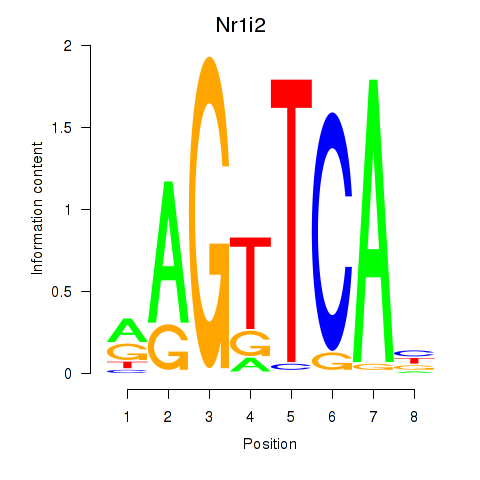
Results for Nr1i2
Z-value: 1.24
Transcription factors associated with Nr1i2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1i2
|
ENSMUSG00000022809.4 | nuclear receptor subfamily 1, group I, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1i2 | mm10_v2_chr16_-_38294774_38294838 | -0.26 | 1.2e-01 | Click! |
Activity profile of Nr1i2 motif
Sorted Z-values of Nr1i2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60582152 | 10.61 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr4_-_61674094 | 10.32 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_60421933 | 9.95 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_61303998 | 9.93 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_62087261 | 9.88 |
ENSMUST00000107488.3
ENSMUST00000107472.1 ENSMUST00000084531.4 |
Mup3
|
major urinary protein 3 |
| chr4_-_60222580 | 9.45 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_61303802 | 9.40 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr4_-_60662358 | 7.66 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr4_-_60139857 | 7.09 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr4_-_61835185 | 6.07 |
ENSMUST00000082287.2
|
Mup5
|
major urinary protein 5 |
| chr19_+_37697792 | 5.60 |
ENSMUST00000025946.5
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr5_-_86926521 | 4.70 |
ENSMUST00000031183.2
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr17_-_32917320 | 4.63 |
ENSMUST00000179434.1
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr17_-_32917048 | 4.58 |
ENSMUST00000054174.7
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr2_-_25469742 | 4.42 |
ENSMUST00000114259.2
ENSMUST00000015234.6 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr10_-_24101951 | 4.08 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr4_-_59960659 | 3.58 |
ENSMUST00000075973.2
|
Mup4
|
major urinary protein 4 |
| chr3_+_129532386 | 3.44 |
ENSMUST00000071402.2
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr6_-_138079916 | 3.16 |
ENSMUST00000171804.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr5_-_87254804 | 3.13 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr1_+_166254095 | 2.91 |
ENSMUST00000111416.1
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr1_-_139781236 | 2.74 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr5_-_77115145 | 2.72 |
ENSMUST00000081964.5
|
Hopx
|
HOP homeobox |
| chr1_+_127729405 | 2.72 |
ENSMUST00000038006.6
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr14_-_34355383 | 2.70 |
ENSMUST00000052126.5
|
Fam25c
|
family with sequence similarity 25, member C |
| chr7_+_27119909 | 2.64 |
ENSMUST00000003100.8
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr16_+_43363855 | 2.55 |
ENSMUST00000156367.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr11_-_113709520 | 2.33 |
ENSMUST00000173655.1
ENSMUST00000100248.4 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr6_+_90333270 | 2.23 |
ENSMUST00000164761.1
ENSMUST00000046128.9 |
Uroc1
|
urocanase domain containing 1 |
| chr1_-_130661613 | 2.19 |
ENSMUST00000027657.7
|
C4bp
|
complement component 4 binding protein |
| chr14_+_52824340 | 2.15 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr1_-_130661584 | 2.08 |
ENSMUST00000137276.2
|
C4bp
|
complement component 4 binding protein |
| chr9_-_98955302 | 2.04 |
ENSMUST00000181706.1
|
Foxl2os
|
forkhead box L2 opposite strand transcript |
| chr4_-_134372529 | 2.02 |
ENSMUST00000030643.2
|
Extl1
|
exostoses (multiple)-like 1 |
| chr17_-_78985428 | 1.99 |
ENSMUST00000118991.1
|
Prkd3
|
protein kinase D3 |
| chr16_+_11406618 | 1.99 |
ENSMUST00000122168.1
|
Snx29
|
sorting nexin 29 |
| chr11_-_3722189 | 1.90 |
ENSMUST00000102950.3
ENSMUST00000101632.3 |
Osbp2
|
oxysterol binding protein 2 |
| chr17_+_21691860 | 1.89 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
| chr1_-_139560158 | 1.86 |
ENSMUST00000160423.1
ENSMUST00000023965.5 |
Cfhr1
|
complement factor H-related 1 |
| chr8_+_76902277 | 1.82 |
ENSMUST00000109912.1
ENSMUST00000128862.1 ENSMUST00000109911.1 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr2_+_71981184 | 1.80 |
ENSMUST00000090826.5
ENSMUST00000102698.3 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr10_+_96136603 | 1.78 |
ENSMUST00000074615.6
|
Gm5426
|
predicted pseudogene 5426 |
| chr7_+_101394361 | 1.77 |
ENSMUST00000154239.1
ENSMUST00000098243.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr11_-_70514608 | 1.72 |
ENSMUST00000021179.3
|
Vmo1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr14_-_118052235 | 1.68 |
ENSMUST00000022725.2
|
Dct
|
dopachrome tautomerase |
| chr2_+_177904285 | 1.64 |
ENSMUST00000099001.3
|
Gm14327
|
predicted gene 14327 |
| chr8_+_119437118 | 1.63 |
ENSMUST00000152420.1
ENSMUST00000098365.3 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr16_+_91269759 | 1.62 |
ENSMUST00000056882.5
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr1_+_88166004 | 1.61 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr18_-_39490649 | 1.59 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr15_+_99393610 | 1.58 |
ENSMUST00000159531.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr1_-_183345296 | 1.57 |
ENSMUST00000109158.3
|
Mia3
|
melanoma inhibitory activity 3 |
| chr5_-_66004278 | 1.55 |
ENSMUST00000067737.5
|
9130230L23Rik
|
RIKEN cDNA 9130230L23 gene |
| chr8_-_72435043 | 1.50 |
ENSMUST00000109974.1
|
Calr3
|
calreticulin 3 |
| chr15_+_99393574 | 1.49 |
ENSMUST00000162624.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr2_+_162987330 | 1.49 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr18_+_20247340 | 1.48 |
ENSMUST00000054128.6
|
Dsg1c
|
desmoglein 1 gamma |
| chr2_+_162987502 | 1.47 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr10_+_128194446 | 1.47 |
ENSMUST00000044776.6
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr6_-_30304513 | 1.46 |
ENSMUST00000094543.2
ENSMUST00000102993.3 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr2_-_25501717 | 1.45 |
ENSMUST00000015227.3
|
C8g
|
complement component 8, gamma polypeptide |
| chr4_+_42735545 | 1.42 |
ENSMUST00000068158.3
|
4930578G10Rik
|
RIKEN cDNA 4930578G10 gene |
| chr7_-_110061319 | 1.40 |
ENSMUST00000098110.2
|
AA474408
|
expressed sequence AA474408 |
| chr2_+_109917639 | 1.39 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr7_+_121707189 | 1.36 |
ENSMUST00000065310.2
|
1700069B07Rik
|
RIKEN cDNA 1700069B07 gene |
| chr13_-_71963713 | 1.34 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr11_-_74925925 | 1.34 |
ENSMUST00000121738.1
|
Srr
|
serine racemase |
| chr6_-_97205549 | 1.33 |
ENSMUST00000164744.1
ENSMUST00000089287.5 |
Uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr1_+_131797381 | 1.33 |
ENSMUST00000112393.2
ENSMUST00000048660.5 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr15_+_7129557 | 1.33 |
ENSMUST00000067190.5
ENSMUST00000164529.1 |
Lifr
|
leukemia inhibitory factor receptor |
| chr17_-_46438471 | 1.32 |
ENSMUST00000087012.5
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr14_+_17981633 | 1.31 |
ENSMUST00000022304.8
|
Thrb
|
thyroid hormone receptor beta |
| chr17_+_8165501 | 1.30 |
ENSMUST00000097419.3
ENSMUST00000024636.8 |
Fgfr1op
|
Fgfr1 oncogene partner |
| chr3_-_92083132 | 1.29 |
ENSMUST00000058150.6
|
Lor
|
loricrin |
| chr16_+_44173239 | 1.29 |
ENSMUST00000119746.1
|
Gm608
|
predicted gene 608 |
| chr14_-_34522801 | 1.28 |
ENSMUST00000022325.1
|
9230112D13Rik
|
RIKEN cDNA 9230112D13 gene |
| chr9_+_65630552 | 1.28 |
ENSMUST00000055844.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr15_-_98221056 | 1.26 |
ENSMUST00000170618.1
ENSMUST00000141911.1 |
Olfr287
|
olfactory receptor 287 |
| chr3_+_32736990 | 1.24 |
ENSMUST00000127477.1
ENSMUST00000121778.1 ENSMUST00000154257.1 |
Ndufb5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 |
| chr17_+_47436731 | 1.24 |
ENSMUST00000150819.2
|
AI661453
|
expressed sequence AI661453 |
| chr6_+_48593883 | 1.24 |
ENSMUST00000154010.1
ENSMUST00000163452.1 ENSMUST00000118229.1 ENSMUST00000009420.8 |
Repin1
|
replication initiator 1 |
| chr4_+_143349757 | 1.24 |
ENSMUST00000052458.2
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr7_+_24611314 | 1.22 |
ENSMUST00000073325.5
|
Phldb3
|
pleckstrin homology-like domain, family B, member 3 |
| chr19_+_41029275 | 1.22 |
ENSMUST00000051806.4
ENSMUST00000112200.1 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr7_-_139582790 | 1.19 |
ENSMUST00000106095.2
|
Nkx6-2
|
NK6 homeobox 2 |
| chr18_-_61014199 | 1.19 |
ENSMUST00000025520.8
|
Slc6a7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 |
| chr1_-_136960427 | 1.19 |
ENSMUST00000027649.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chrX_-_21089229 | 1.18 |
ENSMUST00000040667.6
|
Zfp300
|
zinc finger protein 300 |
| chr11_+_76902152 | 1.18 |
ENSMUST00000102495.1
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr9_+_21835506 | 1.18 |
ENSMUST00000058777.6
|
Gm6484
|
predicted gene 6484 |
| chr16_+_43364145 | 1.17 |
ENSMUST00000148775.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr19_-_47144174 | 1.17 |
ENSMUST00000111813.1
|
Calhm1
|
calcium homeostasis modulator 1 |
| chr1_-_155527083 | 1.16 |
ENSMUST00000097531.2
|
Gm5532
|
predicted gene 5532 |
| chr4_-_43653542 | 1.16 |
ENSMUST00000084646.4
|
Spag8
|
sperm associated antigen 8 |
| chr2_-_71750083 | 1.16 |
ENSMUST00000180494.1
|
Gm17250
|
predicted gene, 17250 |
| chr16_+_37868383 | 1.16 |
ENSMUST00000078717.6
|
Lrrc58
|
leucine rich repeat containing 58 |
| chr18_-_3309858 | 1.14 |
ENSMUST00000144496.1
ENSMUST00000154715.1 |
Crem
|
cAMP responsive element modulator |
| chr10_-_41579207 | 1.14 |
ENSMUST00000095227.3
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr6_-_124917939 | 1.14 |
ENSMUST00000032216.6
|
Ptms
|
parathymosin |
| chr5_-_106696530 | 1.13 |
ENSMUST00000137285.1
ENSMUST00000124263.1 ENSMUST00000112695.1 ENSMUST00000155495.1 ENSMUST00000135108.1 |
Zfp644
|
zinc finger protein 644 |
| chr10_+_44268328 | 1.13 |
ENSMUST00000039286.4
|
Atg5
|
autophagy related 5 |
| chr2_-_176144697 | 1.13 |
ENSMUST00000178872.1
|
Gm2004
|
predicted gene 2004 |
| chr10_+_128194631 | 1.13 |
ENSMUST00000123291.1
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr7_-_4778141 | 1.12 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr10_+_81574699 | 1.12 |
ENSMUST00000131794.1
ENSMUST00000136341.1 |
Tle2
|
transducin-like enhancer of split 2, homolog of Drosophila E(spl) |
| chr17_+_43952999 | 1.11 |
ENSMUST00000177857.1
|
Rcan2
|
regulator of calcineurin 2 |
| chr17_-_56121946 | 1.10 |
ENSMUST00000041357.7
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr17_+_47436615 | 1.10 |
ENSMUST00000037701.6
|
AI661453
|
expressed sequence AI661453 |
| chr14_+_61607455 | 1.09 |
ENSMUST00000051184.8
|
Kcnrg
|
potassium channel regulator |
| chr18_-_3281036 | 1.08 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr7_-_141010759 | 1.07 |
ENSMUST00000026565.6
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr7_+_114718643 | 1.07 |
ENSMUST00000032902.4
ENSMUST00000182816.1 |
Calcb
|
calcitonin-related polypeptide, beta |
| chr4_+_136462250 | 1.06 |
ENSMUST00000084593.2
|
6030445D17Rik
|
RIKEN cDNA 6030445D17 gene |
| chr6_-_5256226 | 1.06 |
ENSMUST00000125686.1
ENSMUST00000031773.2 |
Pon3
|
paraoxonase 3 |
| chr19_-_11081088 | 1.05 |
ENSMUST00000025636.6
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr9_-_22130598 | 1.04 |
ENSMUST00000115315.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr4_-_96077509 | 1.03 |
ENSMUST00000097973.2
ENSMUST00000030305.6 ENSMUST00000107078.2 |
Cyp2j13
|
cytochrome P450, family 2, subfamily j, polypeptide 13 |
| chr4_+_42735912 | 1.03 |
ENSMUST00000107984.1
|
4930578G10Rik
|
RIKEN cDNA 4930578G10 gene |
| chr6_-_11907419 | 1.02 |
ENSMUST00000031637.5
|
Ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr12_-_57546121 | 1.02 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr1_+_59256906 | 1.02 |
ENSMUST00000160662.1
ENSMUST00000114248.2 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr11_+_49280150 | 1.01 |
ENSMUST00000078932.1
|
Olfr1393
|
olfactory receptor 1393 |
| chr9_+_108339048 | 1.01 |
ENSMUST00000082429.5
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_-_26210553 | 1.00 |
ENSMUST00000101447.3
|
5730522E02Rik
|
RIKEN cDNA 5730522E02 gene |
| chr2_+_177768044 | 0.99 |
ENSMUST00000108942.3
|
Gm14322
|
predicted gene 14322 |
| chr19_-_5845471 | 0.99 |
ENSMUST00000174287.1
ENSMUST00000173672.1 |
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr4_+_124880899 | 0.98 |
ENSMUST00000059343.6
|
Epha10
|
Eph receptor A10 |
| chr6_+_34709442 | 0.98 |
ENSMUST00000115021.1
|
Cald1
|
caldesmon 1 |
| chr3_-_138131356 | 0.97 |
ENSMUST00000029805.8
|
Mttp
|
microsomal triglyceride transfer protein |
| chr4_-_129227883 | 0.97 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr15_+_100353149 | 0.97 |
ENSMUST00000075675.5
ENSMUST00000088142.5 ENSMUST00000176287.1 |
AB099516
Mettl7a2
|
cDNA sequence AB099516 methyltransferase like 7A2 |
| chr6_-_24527546 | 0.97 |
ENSMUST00000118558.1
|
Ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr5_-_136565432 | 0.97 |
ENSMUST00000176172.1
|
Cux1
|
cut-like homeobox 1 |
| chr9_+_31280525 | 0.96 |
ENSMUST00000117389.1
|
Prdm10
|
PR domain containing 10 |
| chr18_-_3309723 | 0.95 |
ENSMUST00000136961.1
ENSMUST00000152108.1 |
Crem
|
cAMP responsive element modulator |
| chr19_-_44069736 | 0.95 |
ENSMUST00000172041.1
ENSMUST00000071698.6 ENSMUST00000112028.3 |
Erlin1
|
ER lipid raft associated 1 |
| chr4_+_148140699 | 0.95 |
ENSMUST00000140049.1
ENSMUST00000105707.1 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr10_-_75797528 | 0.95 |
ENSMUST00000120177.1
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr1_-_157108632 | 0.94 |
ENSMUST00000118207.1
ENSMUST00000027884.6 ENSMUST00000121911.1 |
Tex35
|
testis expressed 35 |
| chr9_+_75037838 | 0.93 |
ENSMUST00000169188.1
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr1_-_140183404 | 0.93 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr17_-_37023349 | 0.92 |
ENSMUST00000102665.4
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr3_+_98013503 | 0.92 |
ENSMUST00000079812.6
|
Notch2
|
notch 2 |
| chr4_-_82705735 | 0.92 |
ENSMUST00000155821.1
|
Nfib
|
nuclear factor I/B |
| chr15_-_35938009 | 0.91 |
ENSMUST00000156915.1
|
Cox6c
|
cytochrome c oxidase subunit VIc |
| chr5_-_151190154 | 0.90 |
ENSMUST00000062015.8
ENSMUST00000110483.2 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr4_-_144721404 | 0.90 |
ENSMUST00000036876.7
|
Gm13178
|
predicted gene 13178 |
| chr14_-_20794009 | 0.89 |
ENSMUST00000100837.3
ENSMUST00000080440.6 ENSMUST00000071816.6 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr16_-_34095983 | 0.89 |
ENSMUST00000114973.1
ENSMUST00000114964.1 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr11_+_53457209 | 0.89 |
ENSMUST00000018531.5
|
Shroom1
|
shroom family member 1 |
| chr2_-_90580578 | 0.88 |
ENSMUST00000168621.2
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr17_+_32659410 | 0.88 |
ENSMUST00000165061.1
|
Cyp4f40
|
cytochrome P450, family 4, subfamily f, polypeptide 40 |
| chr4_-_82505274 | 0.88 |
ENSMUST00000050872.8
ENSMUST00000064770.2 |
Nfib
|
nuclear factor I/B |
| chr3_+_122895072 | 0.88 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr17_+_43953191 | 0.88 |
ENSMUST00000044792.4
|
Rcan2
|
regulator of calcineurin 2 |
| chr5_-_116422858 | 0.88 |
ENSMUST00000036991.4
|
Hspb8
|
heat shock protein 8 |
| chr7_+_128003911 | 0.87 |
ENSMUST00000106248.1
|
Trim72
|
tripartite motif-containing 72 |
| chr2_+_152105722 | 0.87 |
ENSMUST00000099225.2
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr7_+_43712505 | 0.86 |
ENSMUST00000066834.6
|
Klk13
|
kallikrein related-peptidase 13 |
| chr7_-_74554474 | 0.86 |
ENSMUST00000134539.1
ENSMUST00000026897.7 ENSMUST00000098371.2 |
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr5_+_144255223 | 0.86 |
ENSMUST00000056578.6
|
Bri3
|
brain protein I3 |
| chr6_-_59426279 | 0.86 |
ENSMUST00000051065.4
|
Gprin3
|
GPRIN family member 3 |
| chr5_-_106696819 | 0.85 |
ENSMUST00000127434.1
ENSMUST00000112696.1 ENSMUST00000112698.1 |
Zfp644
|
zinc finger protein 644 |
| chr2_+_175283298 | 0.84 |
ENSMUST00000098998.3
|
Gm14440
|
predicted gene 14440 |
| chr6_+_48593927 | 0.83 |
ENSMUST00000135151.1
|
Repin1
|
replication initiator 1 |
| chr2_+_80617045 | 0.83 |
ENSMUST00000028384.4
|
Dusp19
|
dual specificity phosphatase 19 |
| chr1_+_9547948 | 0.83 |
ENSMUST00000144177.1
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr9_+_75410145 | 0.83 |
ENSMUST00000180533.1
ENSMUST00000180574.1 |
4933433G15Rik
|
RIKEN cDNA 4933433G15 gene |
| chr11_-_82871133 | 0.82 |
ENSMUST00000071152.7
ENSMUST00000108173.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr7_-_126897424 | 0.82 |
ENSMUST00000120007.1
|
Tmem219
|
transmembrane protein 219 |
| chr9_+_64235201 | 0.82 |
ENSMUST00000039011.3
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr6_-_28447180 | 0.82 |
ENSMUST00000174194.1
|
Pax4
|
paired box gene 4 |
| chr11_+_51261719 | 0.81 |
ENSMUST00000130641.1
|
Clk4
|
CDC like kinase 4 |
| chr9_+_78051938 | 0.80 |
ENSMUST00000024104.7
|
Gcm1
|
glial cells missing homolog 1 (Drosophila) |
| chr9_-_89092835 | 0.80 |
ENSMUST00000167113.1
|
Trim43b
|
tripartite motif-containing 43B |
| chr6_+_21986887 | 0.80 |
ENSMUST00000151315.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr8_-_13200576 | 0.80 |
ENSMUST00000165605.2
|
Grtp1
|
GH regulated TBC protein 1 |
| chr2_-_136891363 | 0.80 |
ENSMUST00000028730.6
ENSMUST00000110089.2 |
Mkks
|
McKusick-Kaufman syndrome |
| chr5_+_30466044 | 0.80 |
ENSMUST00000031078.3
ENSMUST00000114743.1 |
1700001C02Rik
|
RIKEN cDNA 1700001C02 gene |
| chr14_+_103046977 | 0.79 |
ENSMUST00000022722.6
|
Irg1
|
immunoresponsive gene 1 |
| chr5_-_48889531 | 0.79 |
ENSMUST00000176978.1
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr13_-_73328442 | 0.79 |
ENSMUST00000022097.5
|
Ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr18_-_61036189 | 0.78 |
ENSMUST00000025521.8
|
Cdx1
|
caudal type homeobox 1 |
| chr9_+_40187079 | 0.78 |
ENSMUST00000062229.5
|
Olfr986
|
olfactory receptor 986 |
| chr11_+_98960412 | 0.78 |
ENSMUST00000107473.2
|
Rara
|
retinoic acid receptor, alpha |
| chr1_-_140183283 | 0.78 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr5_+_119834663 | 0.77 |
ENSMUST00000018407.6
|
Tbx5
|
T-box 5 |
| chr14_-_16243309 | 0.77 |
ENSMUST00000112625.1
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr10_-_81600857 | 0.77 |
ENSMUST00000151858.1
ENSMUST00000142948.1 ENSMUST00000072020.2 |
Tle6
|
transducin-like enhancer of split 6, homolog of Drosophila E(spl) |
| chr1_+_6214627 | 0.76 |
ENSMUST00000027040.6
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr6_-_99632376 | 0.76 |
ENSMUST00000176255.1
|
Gm20696
|
predicted gene 20696 |
| chr3_-_10335650 | 0.76 |
ENSMUST00000078748.3
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr7_+_105554360 | 0.75 |
ENSMUST00000046983.8
|
Smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr2_-_175133322 | 0.75 |
ENSMUST00000099029.3
|
Gm14399
|
predicted gene 14399 |
| chr7_-_25390098 | 0.75 |
ENSMUST00000054301.7
|
Lipe
|
lipase, hormone sensitive |
| chr3_-_96220880 | 0.75 |
ENSMUST00000090782.3
|
Hist2h2ac
|
histone cluster 2, H2ac |
| chr10_+_94198955 | 0.75 |
ENSMUST00000020209.9
ENSMUST00000179990.1 |
Ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr8_+_46739745 | 0.74 |
ENSMUST00000034041.7
|
Irf2
|
interferon regulatory factor 2 |
| chr7_+_105650813 | 0.74 |
ENSMUST00000145988.2
|
Dnhd1
|
dynein heavy chain domain 1 |
| chr4_+_80910646 | 0.73 |
ENSMUST00000055922.3
|
Lurap1l
|
leucine rich adaptor protein 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1i2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.4 | 5.6 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.9 | 3.6 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.9 | 4.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.6 | 7.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.6 | 3.1 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.6 | 1.8 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.6 | 1.7 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.5 | 0.5 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.5 | 1.4 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.5 | 1.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.5 | 2.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.4 | 1.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 1.8 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 2.2 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.4 | 2.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 11.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.4 | 3.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 1.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 1.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 1.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.4 | 1.1 | GO:0009804 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.3 | 2.7 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.3 | 1.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.3 | 1.3 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.3 | 1.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.3 | 1.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 1.0 | GO:0010949 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.3 | 0.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 0.9 | GO:1990705 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.3 | 0.9 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.3 | 0.9 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.3 | 1.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.3 | 0.9 | GO:0042374 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.3 | 3.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 2.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 1.3 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.3 | 1.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 1.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.3 | 0.8 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 0.7 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.2 | 1.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 1.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.7 | GO:1900245 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.7 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.2 | 0.6 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.2 | 1.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.6 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 0.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.2 | GO:0015867 | ATP transport(GO:0015867) sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.2 | 2.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 0.6 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 0.2 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 1.7 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 1.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 1.0 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.2 | 1.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 0.3 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.2 | 1.6 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.8 | GO:0060296 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 0.6 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 0.5 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.6 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.1 | 1.0 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.1 | 1.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 | 0.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.8 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 0.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.6 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 1.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 2.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.3 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.7 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.3 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 1.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.6 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 3.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.4 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.1 | 0.9 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.7 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.1 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 2.1 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 1.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 3.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.2 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.8 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.2 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.3 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.1 | 0.2 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 1.3 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 1.5 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 2.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 2.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 1.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.8 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 3.0 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 3.7 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.6 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.9 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.1 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.8 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 3.2 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 1.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 1.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.3 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.3 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 1.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.5 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.0 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 1.0 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.3 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 1.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 1.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.8 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 1.0 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.9 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.3 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 3.0 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.9 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.3 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 5.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 1.5 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.6 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.7 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0003433 | growth plate cartilage chondrocyte differentiation(GO:0003418) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0034205 | beta-amyloid formation(GO:0034205) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.1 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 0.5 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of luteinizing hormone secretion(GO:0033686) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 1.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.8 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 1.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.3 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.3 | GO:0019228 | neuronal action potential(GO:0019228) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.4 | 1.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 1.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.3 | 1.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 0.7 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 1.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.2 | 1.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 1.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 0.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 5.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.4 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 1.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 2.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 6.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 2.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 4.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 4.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 7.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.9 | 5.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.1 | 4.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.7 | 20.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.7 | 2.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.6 | 1.8 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.5 | 1.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.5 | 3.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 1.3 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.4 | 1.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.4 | 2.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 4.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 1.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 1.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.4 | 1.1 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.3 | 2.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 2.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.6 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 0.9 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.3 | 3.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 2.0 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.3 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 9.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 1.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 1.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 1.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 0.6 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.2 | 0.8 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 0.8 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 1.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) NADH binding(GO:0070404) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 3.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.6 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 1.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 2.6 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.3 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 2.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 1.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.5 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 2.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 2.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.2 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 2.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 2.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 6.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 2.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0016893 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters(GO:0016893) |
| 0.0 | 2.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 2.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.9 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 5.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 3.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 2.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 8.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 8.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.5 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |