Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
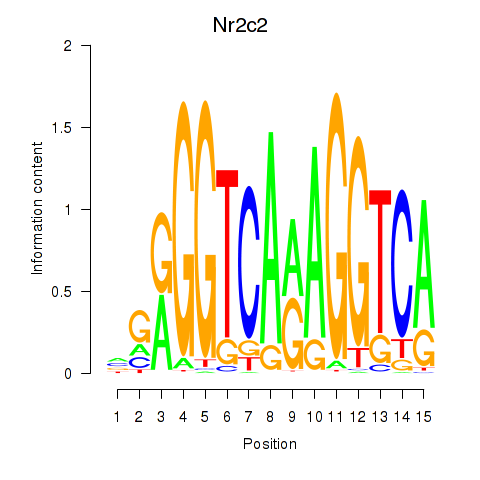
Results for Nr2c2
Z-value: 1.81
Transcription factors associated with Nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c2
|
ENSMUSG00000005893.8 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c2 | mm10_v2_chr6_+_92091378_92091390 | -0.66 | 1.1e-05 | Click! |
Activity profile of Nr2c2 motif
Sorted Z-values of Nr2c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_62087261 | 24.56 |
ENSMUST00000107488.3
ENSMUST00000107472.1 ENSMUST00000084531.4 |
Mup3
|
major urinary protein 3 |
| chr4_-_61303998 | 24.31 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_60501903 | 23.90 |
ENSMUST00000084548.4
ENSMUST00000103012.3 ENSMUST00000107499.3 |
Mup1
|
major urinary protein 1 |
| chr4_-_61303802 | 23.18 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr4_-_62054112 | 22.71 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr4_-_60741275 | 22.66 |
ENSMUST00000117932.1
|
Mup12
|
major urinary protein 12 |
| chr19_+_46131888 | 19.81 |
ENSMUST00000043739.3
|
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr7_-_99695628 | 16.69 |
ENSMUST00000145381.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr7_-_99695809 | 16.12 |
ENSMUST00000107086.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr7_-_99695572 | 15.54 |
ENSMUST00000137914.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr15_-_82764176 | 13.74 |
ENSMUST00000055721.4
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr3_+_138415484 | 12.55 |
ENSMUST00000161312.1
ENSMUST00000013458.8 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr3_+_138374121 | 9.17 |
ENSMUST00000171054.1
|
Adh6-ps1
|
alcohol dehydrogenase 6 (class V), pseudogene 1 |
| chr9_-_86695897 | 8.41 |
ENSMUST00000034989.8
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr9_-_46235631 | 7.91 |
ENSMUST00000118649.1
|
Apoc3
|
apolipoprotein C-III |
| chr7_+_44207307 | 7.90 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr11_-_78422217 | 7.45 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr5_+_114146525 | 7.31 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr7_-_14438538 | 7.27 |
ENSMUST00000168252.2
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr9_-_22002599 | 6.70 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr1_+_74713551 | 6.65 |
ENSMUST00000027356.5
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr8_+_104591464 | 6.17 |
ENSMUST00000059588.6
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr10_+_84756055 | 6.07 |
ENSMUST00000060397.6
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr1_-_120120138 | 5.79 |
ENSMUST00000112648.1
ENSMUST00000128408.1 |
Dbi
|
diazepam binding inhibitor |
| chr6_+_72636244 | 5.62 |
ENSMUST00000101278.2
|
Gm15401
|
predicted gene 15401 |
| chr1_+_166254095 | 5.50 |
ENSMUST00000111416.1
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr3_+_63295815 | 5.26 |
ENSMUST00000029400.1
|
Mme
|
membrane metallo endopeptidase |
| chr18_-_61911783 | 5.17 |
ENSMUST00000049378.8
ENSMUST00000166783.1 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr15_+_7129557 | 5.12 |
ENSMUST00000067190.5
ENSMUST00000164529.1 |
Lifr
|
leukemia inhibitory factor receptor |
| chr8_-_93197799 | 5.10 |
ENSMUST00000034172.7
|
Ces1d
|
carboxylesterase 1D |
| chr19_+_36554661 | 5.00 |
ENSMUST00000169036.2
ENSMUST00000047247.5 |
Hectd2
|
HECT domain containing 2 |
| chr1_+_195017399 | 4.98 |
ENSMUST00000181273.1
|
A330023F24Rik
|
RIKEN cDNA A330023F24 gene |
| chr14_+_27622433 | 4.56 |
ENSMUST00000090302.5
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr6_-_85707858 | 4.41 |
ENSMUST00000179613.1
|
Gm4477
|
predicted gene 4477 |
| chr17_-_83846769 | 4.39 |
ENSMUST00000000687.7
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr4_-_134372529 | 4.19 |
ENSMUST00000030643.2
|
Extl1
|
exostoses (multiple)-like 1 |
| chr3_+_94342092 | 4.11 |
ENSMUST00000029794.5
|
Them5
|
thioesterase superfamily member 5 |
| chr3_-_129332713 | 4.09 |
ENSMUST00000029658.7
|
Enpep
|
glutamyl aminopeptidase |
| chr8_+_94525067 | 3.94 |
ENSMUST00000098489.4
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr7_+_44465391 | 3.93 |
ENSMUST00000035929.4
ENSMUST00000146128.1 |
Aspdh
|
aspartate dehydrogenase domain containing |
| chr4_+_100478806 | 3.91 |
ENSMUST00000133493.2
ENSMUST00000092730.3 ENSMUST00000106979.3 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr19_+_12633507 | 3.87 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr5_-_135744206 | 3.80 |
ENSMUST00000153399.1
ENSMUST00000043378.2 |
Tmem120a
|
transmembrane protein 120A |
| chr5_+_130448801 | 3.75 |
ENSMUST00000111288.2
|
Caln1
|
calneuron 1 |
| chr4_+_148602527 | 3.70 |
ENSMUST00000105701.2
ENSMUST00000052060.6 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr8_-_71537402 | 3.67 |
ENSMUST00000051672.7
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr4_+_97777780 | 3.66 |
ENSMUST00000107062.2
ENSMUST00000052018.5 ENSMUST00000107057.1 |
Nfia
|
nuclear factor I/A |
| chr16_+_5007283 | 3.64 |
ENSMUST00000184439.1
|
Smim22
|
small integral membrane protein 22 |
| chr8_-_70632419 | 3.62 |
ENSMUST00000110103.1
|
Gdf15
|
growth differentiation factor 15 |
| chr10_-_89506631 | 3.61 |
ENSMUST00000058126.8
ENSMUST00000105296.2 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr19_+_12633303 | 3.58 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr6_+_108828633 | 3.49 |
ENSMUST00000089162.3
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr13_+_64248649 | 3.38 |
ENSMUST00000181403.1
|
1810034E14Rik
|
RIKEN cDNA 1810034E14 gene |
| chr8_+_13895816 | 3.37 |
ENSMUST00000084055.7
|
Gm7676
|
predicted gene 7676 |
| chr15_+_100304782 | 3.36 |
ENSMUST00000067752.3
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr4_+_97777606 | 3.35 |
ENSMUST00000075448.6
ENSMUST00000092532.6 |
Nfia
|
nuclear factor I/A |
| chr11_+_102761402 | 3.32 |
ENSMUST00000103081.4
ENSMUST00000068150.5 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr7_-_68275098 | 3.32 |
ENSMUST00000135564.1
|
Gm16157
|
predicted gene 16157 |
| chr2_-_25356319 | 3.30 |
ENSMUST00000028332.7
|
Dpp7
|
dipeptidylpeptidase 7 |
| chr5_-_136567242 | 3.27 |
ENSMUST00000175975.2
ENSMUST00000176216.2 ENSMUST00000176745.1 |
Cux1
|
cut-like homeobox 1 |
| chr12_+_108792946 | 3.27 |
ENSMUST00000021692.7
|
Yy1
|
YY1 transcription factor |
| chr1_+_74284930 | 3.25 |
ENSMUST00000113805.1
ENSMUST00000027370.6 ENSMUST00000087226.4 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chrX_-_100412587 | 3.25 |
ENSMUST00000033567.8
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr19_+_46707443 | 3.25 |
ENSMUST00000003655.7
|
As3mt
|
arsenic (+3 oxidation state) methyltransferase |
| chr9_-_65885024 | 3.23 |
ENSMUST00000122410.1
ENSMUST00000117083.1 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr10_-_79983970 | 3.22 |
ENSMUST00000124536.1
|
Tmem259
|
transmembrane protein 259 |
| chr8_-_91801948 | 3.14 |
ENSMUST00000175795.1
|
Irx3
|
Iroquois related homeobox 3 (Drosophila) |
| chr10_-_75773350 | 3.13 |
ENSMUST00000001716.7
|
Ddt
|
D-dopachrome tautomerase |
| chr1_-_120120937 | 3.13 |
ENSMUST00000151708.1
|
Dbi
|
diazepam binding inhibitor |
| chr10_-_79984227 | 3.09 |
ENSMUST00000052885.7
|
Tmem259
|
transmembrane protein 259 |
| chr17_-_34028044 | 3.09 |
ENSMUST00000045467.7
ENSMUST00000114303.3 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr11_+_98348404 | 3.08 |
ENSMUST00000078694.6
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr10_-_41611319 | 3.08 |
ENSMUST00000179614.1
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr7_-_105600103 | 3.07 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr9_+_20868628 | 3.07 |
ENSMUST00000043911.7
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene |
| chr7_+_30232310 | 3.05 |
ENSMUST00000108193.1
ENSMUST00000108192.1 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr9_-_50746501 | 3.03 |
ENSMUST00000034564.1
|
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr15_-_77533312 | 3.01 |
ENSMUST00000062562.5
|
Apol7c
|
apolipoprotein L 7c |
| chr15_-_76009440 | 2.99 |
ENSMUST00000170153.1
|
Fam83h
|
family with sequence similarity 83, member H |
| chr6_-_95718800 | 2.95 |
ENSMUST00000079847.5
|
Suclg2
|
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
| chr4_+_41760454 | 2.92 |
ENSMUST00000108040.1
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr9_+_107576915 | 2.92 |
ENSMUST00000112387.2
ENSMUST00000123005.1 ENSMUST00000010195.7 ENSMUST00000144392.1 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr8_-_86580664 | 2.92 |
ENSMUST00000131423.1
ENSMUST00000152438.1 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr4_-_41640322 | 2.91 |
ENSMUST00000127306.1
|
Enho
|
energy homeostasis associated |
| chr17_+_35470083 | 2.89 |
ENSMUST00000174525.1
ENSMUST00000068291.6 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr4_-_103215147 | 2.88 |
ENSMUST00000150285.1
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr3_+_135825788 | 2.84 |
ENSMUST00000167390.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr2_-_148045891 | 2.83 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr7_+_64501687 | 2.83 |
ENSMUST00000032732.8
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr7_-_63212514 | 2.82 |
ENSMUST00000032738.5
|
Chrna7
|
cholinergic receptor, nicotinic, alpha polypeptide 7 |
| chr8_-_91801547 | 2.78 |
ENSMUST00000093312.4
|
Irx3
|
Iroquois related homeobox 3 (Drosophila) |
| chr16_+_5007306 | 2.78 |
ENSMUST00000178155.2
ENSMUST00000184256.1 ENSMUST00000185147.1 |
Smim22
|
small integral membrane protein 22 |
| chr11_-_116198701 | 2.77 |
ENSMUST00000072948.4
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr17_-_73950172 | 2.76 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr2_-_25500613 | 2.76 |
ENSMUST00000040042.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr19_-_42202150 | 2.76 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr15_-_76090013 | 2.75 |
ENSMUST00000019516.4
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr8_+_36457548 | 2.74 |
ENSMUST00000135373.1
ENSMUST00000147525.1 |
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr5_-_139814231 | 2.74 |
ENSMUST00000044002.4
|
Tmem184a
|
transmembrane protein 184a |
| chr1_+_171225054 | 2.70 |
ENSMUST00000111321.1
ENSMUST00000005824.5 ENSMUST00000111320.1 ENSMUST00000111319.1 |
Apoa2
|
apolipoprotein A-II |
| chr2_+_162987502 | 2.68 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr17_-_56717681 | 2.68 |
ENSMUST00000164907.1
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr2_-_84743655 | 2.68 |
ENSMUST00000181711.1
|
Gm19426
|
predicted gene, 19426 |
| chr17_-_56005566 | 2.64 |
ENSMUST00000043785.6
|
Stap2
|
signal transducing adaptor family member 2 |
| chr12_-_103830373 | 2.62 |
ENSMUST00000164454.2
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr3_+_62338344 | 2.62 |
ENSMUST00000079300.6
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr17_-_57247632 | 2.60 |
ENSMUST00000005975.6
|
Gpr108
|
G protein-coupled receptor 108 |
| chr12_-_79192248 | 2.59 |
ENSMUST00000161204.1
|
Rdh11
|
retinol dehydrogenase 11 |
| chrX_-_139782353 | 2.57 |
ENSMUST00000101217.3
|
Ripply1
|
ripply1 homolog (zebrafish) |
| chr8_-_121578755 | 2.57 |
ENSMUST00000181663.1
ENSMUST00000059018.7 |
Fbxo31
|
F-box protein 31 |
| chr6_-_29507946 | 2.57 |
ENSMUST00000101614.3
ENSMUST00000078112.6 |
Kcp
|
kielin/chordin-like protein |
| chr19_-_3912711 | 2.56 |
ENSMUST00000075092.6
|
Ndufs8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8 |
| chr17_-_35909626 | 2.54 |
ENSMUST00000141132.1
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr11_-_116199040 | 2.53 |
ENSMUST00000066587.5
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr5_-_139819906 | 2.46 |
ENSMUST00000147328.1
|
Tmem184a
|
transmembrane protein 184a |
| chr4_+_118428078 | 2.45 |
ENSMUST00000006557.6
ENSMUST00000167636.1 ENSMUST00000102673.4 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr2_+_162987330 | 2.45 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr5_+_92571544 | 2.40 |
ENSMUST00000082382.7
|
Fam47e
|
family with sequence similarity 47, member E |
| chr11_+_69991633 | 2.39 |
ENSMUST00000108592.1
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr11_-_106613370 | 2.38 |
ENSMUST00000128933.1
|
Tex2
|
testis expressed gene 2 |
| chr3_+_135826075 | 2.37 |
ENSMUST00000029810.5
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr10_-_42478280 | 2.36 |
ENSMUST00000151747.1
|
Lace1
|
lactation elevated 1 |
| chr3_-_88334428 | 2.36 |
ENSMUST00000107552.1
|
Tmem79
|
transmembrane protein 79 |
| chr7_-_31055594 | 2.35 |
ENSMUST00000039909.6
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr17_+_26113286 | 2.35 |
ENSMUST00000025010.7
|
Tmem8
|
transmembrane protein 8 (five membrane-spanning domains) |
| chr4_+_85205417 | 2.34 |
ENSMUST00000030212.8
ENSMUST00000107189.1 ENSMUST00000107184.1 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr5_+_92571477 | 2.33 |
ENSMUST00000176621.1
ENSMUST00000175974.1 ENSMUST00000131166.2 ENSMUST00000176448.1 |
Fam47e
|
family with sequence similarity 47, member E |
| chr16_-_38294774 | 2.31 |
ENSMUST00000023504.4
|
Nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr8_-_72435043 | 2.29 |
ENSMUST00000109974.1
|
Calr3
|
calreticulin 3 |
| chr7_+_3617357 | 2.27 |
ENSMUST00000076657.4
ENSMUST00000108644.1 |
Ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr8_+_105708270 | 2.25 |
ENSMUST00000013302.5
|
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr8_-_110039330 | 2.23 |
ENSMUST00000109222.2
|
Chst4
|
carbohydrate (chondroitin 6/keratan) sulfotransferase 4 |
| chr19_-_44107447 | 2.23 |
ENSMUST00000119591.1
ENSMUST00000026217.4 |
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chrX_+_101376359 | 2.20 |
ENSMUST00000119080.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr3_-_57692537 | 2.20 |
ENSMUST00000099091.3
|
Gm410
|
predicted gene 410 |
| chr16_-_90934506 | 2.19 |
ENSMUST00000142340.1
|
1110004E09Rik
|
RIKEN cDNA 1110004E09 gene |
| chr10_-_42478488 | 2.19 |
ENSMUST00000041024.8
|
Lace1
|
lactation elevated 1 |
| chr2_-_164473714 | 2.16 |
ENSMUST00000017864.2
|
Trp53tg5
|
transformation related protein 53 target 5 |
| chr5_+_53267103 | 2.15 |
ENSMUST00000121042.1
|
Smim20
|
small integral membrane protein 20 |
| chr16_-_93929512 | 2.14 |
ENSMUST00000177648.1
|
Cldn14
|
claudin 14 |
| chr19_-_4498574 | 2.13 |
ENSMUST00000048482.6
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr15_+_88819584 | 2.13 |
ENSMUST00000024042.3
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr5_+_113772748 | 2.12 |
ENSMUST00000026937.5
ENSMUST00000112311.1 ENSMUST00000112312.1 |
Iscu
|
IscU iron-sulfur cluster scaffold homolog (E. coli) |
| chr2_-_27246814 | 2.11 |
ENSMUST00000149733.1
|
Sardh
|
sarcosine dehydrogenase |
| chr2_+_34772089 | 2.11 |
ENSMUST00000028222.6
ENSMUST00000100171.2 |
Hspa5
|
heat shock protein 5 |
| chr18_-_36744518 | 2.10 |
ENSMUST00000014438.4
|
Ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr17_+_56717759 | 2.08 |
ENSMUST00000002452.6
|
Ndufa11
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 11 |
| chr17_+_35262730 | 2.07 |
ENSMUST00000172785.1
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr11_+_102041509 | 2.07 |
ENSMUST00000123895.1
ENSMUST00000017453.5 ENSMUST00000107163.2 ENSMUST00000107164.2 |
Cd300lg
|
CD300 antigen like family member G |
| chr15_-_74728011 | 2.04 |
ENSMUST00000023261.2
|
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr9_-_22135675 | 2.02 |
ENSMUST00000165735.1
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr8_+_13026024 | 2.02 |
ENSMUST00000033820.3
|
F7
|
coagulation factor VII |
| chr2_+_32609043 | 2.02 |
ENSMUST00000128811.1
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr9_-_78322357 | 2.00 |
ENSMUST00000095071.4
|
Gm8074
|
predicted gene 8074 |
| chr2_+_4718145 | 2.00 |
ENSMUST00000056914.6
|
Bend7
|
BEN domain containing 7 |
| chr19_-_6921804 | 1.99 |
ENSMUST00000025906.4
|
Esrra
|
estrogen related receptor, alpha |
| chr16_+_3884657 | 1.98 |
ENSMUST00000176625.1
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr2_+_132263136 | 1.98 |
ENSMUST00000110158.1
ENSMUST00000103181.4 |
Cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr3_+_89136133 | 1.96 |
ENSMUST00000047111.6
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr9_+_119402444 | 1.96 |
ENSMUST00000035093.8
ENSMUST00000165044.1 |
Acvr2b
|
activin receptor IIB |
| chr4_+_118429701 | 1.95 |
ENSMUST00000067896.3
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr11_+_103103051 | 1.93 |
ENSMUST00000152971.1
|
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr6_+_83054653 | 1.93 |
ENSMUST00000092618.6
|
Aup1
|
ancient ubiquitous protein 1 |
| chr4_+_11156411 | 1.93 |
ENSMUST00000029865.3
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1 |
| chr13_-_29855630 | 1.92 |
ENSMUST00000091674.5
ENSMUST00000006353.7 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr11_+_70519183 | 1.92 |
ENSMUST00000057685.2
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr2_+_119167758 | 1.92 |
ENSMUST00000057454.3
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr13_+_91741507 | 1.92 |
ENSMUST00000022120.4
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr13_-_29984219 | 1.91 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chr3_-_88334482 | 1.90 |
ENSMUST00000001456.4
|
Tmem79
|
transmembrane protein 79 |
| chr2_-_160872829 | 1.90 |
ENSMUST00000176141.1
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr10_-_127621107 | 1.89 |
ENSMUST00000049149.8
|
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr6_+_83156401 | 1.88 |
ENSMUST00000032106.4
|
1700003E16Rik
|
RIKEN cDNA 1700003E16 gene |
| chr15_-_27630644 | 1.86 |
ENSMUST00000059662.7
|
Fam105b
|
family with sequence similarity 105, member B |
| chr11_-_77894096 | 1.86 |
ENSMUST00000017597.4
|
Pipox
|
pipecolic acid oxidase |
| chr10_+_128790903 | 1.85 |
ENSMUST00000026411.6
|
Mmp19
|
matrix metallopeptidase 19 |
| chr10_-_127666598 | 1.84 |
ENSMUST00000099157.3
|
Nab2
|
Ngfi-A binding protein 2 |
| chr17_+_35424842 | 1.84 |
ENSMUST00000174699.1
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chrX_-_74023745 | 1.84 |
ENSMUST00000114353.3
ENSMUST00000101458.2 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr8_-_84846860 | 1.84 |
ENSMUST00000003912.6
|
Calr
|
calreticulin |
| chr7_-_30232186 | 1.83 |
ENSMUST00000006254.5
|
Tbcb
|
tubulin folding cofactor B |
| chr15_-_84123174 | 1.83 |
ENSMUST00000019012.3
|
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr2_+_69790968 | 1.83 |
ENSMUST00000180290.1
|
Phospho2
|
phosphatase, orphan 2 |
| chr9_+_65630552 | 1.83 |
ENSMUST00000055844.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr18_+_74779190 | 1.82 |
ENSMUST00000041053.9
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chr10_+_76575640 | 1.82 |
ENSMUST00000001183.7
|
Ftcd
|
formiminotransferase cyclodeaminase |
| chr9_-_44417983 | 1.81 |
ENSMUST00000053286.7
|
Ccdc84
|
coiled-coil domain containing 84 |
| chr10_+_43524080 | 1.81 |
ENSMUST00000057649.6
|
Gm9803
|
predicted gene 9803 |
| chr10_-_75822521 | 1.80 |
ENSMUST00000160211.1
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr15_+_3270767 | 1.80 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr2_+_25557847 | 1.79 |
ENSMUST00000015236.3
|
Edf1
|
endothelial differentiation-related factor 1 |
| chr11_+_97798995 | 1.78 |
ENSMUST00000143571.1
|
Lasp1
|
LIM and SH3 protein 1 |
| chr16_-_17928136 | 1.77 |
ENSMUST00000003622.8
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr7_+_30232032 | 1.77 |
ENSMUST00000149654.1
|
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr5_+_30232581 | 1.76 |
ENSMUST00000145167.1
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr3_+_89136572 | 1.76 |
ENSMUST00000107482.3
ENSMUST00000127058.1 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr7_+_43444104 | 1.74 |
ENSMUST00000004729.3
|
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr17_-_29716969 | 1.74 |
ENSMUST00000129091.1
ENSMUST00000128751.1 |
Ccdc167
|
coiled-coil domain containing 167 |
| chr19_-_21472552 | 1.73 |
ENSMUST00000087600.3
|
Gda
|
guanine deaminase |
| chr6_-_87690819 | 1.73 |
ENSMUST00000162547.1
|
1810020O05Rik
|
Riken cDNA 1810020O05 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 48.4 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 3.8 | 22.7 | GO:0008355 | olfactory learning(GO:0008355) |
| 3.1 | 12.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 2.4 | 7.3 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 2.2 | 23.9 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 2.0 | 24.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 2.0 | 7.9 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.8 | 8.9 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 1.8 | 5.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.7 | 5.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.6 | 4.8 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 1.4 | 4.3 | GO:0042335 | cuticle development(GO:0042335) |
| 1.2 | 3.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 1.2 | 6.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 1.2 | 3.6 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 1.2 | 8.4 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 1.2 | 3.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.1 | 3.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 1.0 | 5.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.0 | 3.9 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.0 | 3.9 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 1.0 | 3.9 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.0 | 2.9 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 2.8 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.9 | 2.8 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.9 | 2.8 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.9 | 24.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.9 | 5.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.9 | 5.2 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.8 | 5.9 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.8 | 7.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.8 | 3.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.7 | 2.2 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.7 | 3.7 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.7 | 7.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.7 | 2.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.7 | 2.1 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.7 | 2.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.7 | 2.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.7 | 4.6 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.6 | 2.6 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.6 | 1.9 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.6 | 2.5 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.6 | 1.9 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.6 | 3.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.6 | 1.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.6 | 3.0 | GO:1901098 | negative regulation of autophagosome maturation(GO:1901097) positive regulation of autophagosome maturation(GO:1901098) |
| 0.6 | 4.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.6 | 4.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.6 | 1.7 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.6 | 1.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.6 | 2.8 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.6 | 1.7 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.5 | 4.4 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.5 | 3.3 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.5 | 1.6 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.5 | 4.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.5 | 1.5 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.5 | 2.0 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.5 | 1.5 | GO:0090155 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.5 | 2.4 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.5 | 1.4 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.5 | 2.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.5 | 3.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.5 | 1.8 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.5 | 3.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.5 | 6.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.4 | 6.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 1.7 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.4 | 2.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.4 | 1.6 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.4 | 1.2 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.4 | 1.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.4 | 1.2 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.4 | 1.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.4 | 1.5 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.4 | 3.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 1.8 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.4 | 1.8 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.4 | 2.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 6.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.3 | 2.4 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 3.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 1.7 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.3 | 1.4 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.3 | 1.0 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 3.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.3 | 4.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.3 | 0.6 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.3 | 1.9 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 2.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.3 | 2.9 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.3 | 1.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 0.6 | GO:0034442 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.3 | 2.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 12.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 | 2.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 3.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 0.6 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.3 | 2.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 1.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 3.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 0.8 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.3 | 1.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.3 | 2.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.8 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.3 | 1.8 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.3 | 1.3 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 0.8 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.2 | 1.5 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.2 | 2.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 0.7 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.7 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.2 | 0.7 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.2 | 7.9 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 1.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 4.0 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 2.2 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.2 | 2.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 0.9 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.2 | 2.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 1.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 13.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 0.8 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 1.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 1.8 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 7.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 2.9 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 3.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 2.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 1.7 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.2 | 2.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.5 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 1.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 2.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 2.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 0.5 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 1.8 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 0.5 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.2 | 0.9 | GO:0070627 | ferrous iron import(GO:0070627) |
| 0.2 | 2.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.6 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.6 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.7 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 1.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.7 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.1 | 0.6 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 1.3 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 2.4 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.1 | 1.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 2.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 2.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.8 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 1.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 2.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.9 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.2 | GO:0072141 | mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.1 | 1.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 4.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.9 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.7 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.5 | GO:0032819 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 1.4 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 0.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 7.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 3.4 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 0.2 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 | 1.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 1.0 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.9 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.1 | 0.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 1.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.9 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 2.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 3.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 2.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.2 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 1.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.8 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 2.4 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.6 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.2 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 2.0 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 3.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 6.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.8 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.8 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 1.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.8 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.4 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.9 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 1.3 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 5.1 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 1.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 3.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.8 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 1.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 2.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 3.2 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:0036019 | endolysosome(GO:0036019) |
| 0.9 | 2.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.7 | 8.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.7 | 8.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 2.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.6 | 0.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.6 | 3.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.5 | 48.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 3.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.4 | 3.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 1.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 1.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 1.8 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 2.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.3 | 1.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 8.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 2.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 14.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 1.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 2.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.3 | 2.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 0.8 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.3 | 2.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 2.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 3.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 5.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 8.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 3.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 31.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 1.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 4.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 6.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 2.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.6 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.2 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.7 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 2.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 11.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 5.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 5.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.8 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 18.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 7.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 5.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 17.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 5.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 7.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 3.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 6.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 29.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0000138 | Golgi cis cisterna(GO:0000137) Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 21.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 4.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 46.6 | GO:0005186 | pheromone activity(GO:0005186) |
| 4.2 | 12.5 | GO:0004024 | NADPH:quinone reductase activity(GO:0003960) alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 3.0 | 8.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 3.0 | 11.8 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.8 | 8.4 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 2.3 | 48.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 2.0 | 24.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 2.0 | 9.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.9 | 7.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.7 | 5.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 1.7 | 5.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.5 | 6.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.5 | 2.9 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 1.4 | 4.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 1.3 | 3.9 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 1.2 | 3.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.1 | 3.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 1.0 | 3.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) D5 dopamine receptor binding(GO:0031752) |
| 1.0 | 24.6 | GO:0005550 | pheromone binding(GO:0005550) |
| 1.0 | 2.9 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.9 | 5.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.8 | 2.4 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.8 | 3.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.7 | 2.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 3.6 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.7 | 2.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.7 | 2.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.6 | 3.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.6 | 3.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 1.7 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.5 | 3.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.5 | 6.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.5 | 2.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.5 | 1.5 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.5 | 1.5 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.5 | 1.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 2.8 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.5 | 2.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 1.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.4 | 2.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.4 | 1.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.4 | 1.2 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.4 | 1.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 3.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 1.9 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.4 | 2.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 2.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.4 | 3.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 3.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 1.8 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.3 | 4.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 7.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 4.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 3.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 2.9 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 20.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 2.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 1.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.3 | 8.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 1.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.3 | 1.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 5.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 1.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 1.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.3 | 1.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 1.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 3.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 7.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 5.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 3.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.2 | 2.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.6 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 4.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 6.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 0.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.9 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 0.8 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 1.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.7 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.1 | 3.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 1.8 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 2.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 2.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.3 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 2.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 4.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.5 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 1.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 4.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 3.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 4.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.5 | GO:0017002 | activin receptor activity, type I(GO:0016361) activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.9 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 7.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 6.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 7.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 3.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 3.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 7.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 2.5 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 3.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 2.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 4.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 8.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 5.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 4.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 44.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 2.1 | 12.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.9 | 25.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.6 | 4.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.6 | 7.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 6.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 7.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 6.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 3.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 5.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.4 | 5.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.4 | 10.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 4.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 5.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 6.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 2.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 4.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 2.8 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 13.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 5.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 6.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 1.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 4.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 10.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 3.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 6.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 2.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |