Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr4a2
Z-value: 0.65
Transcription factors associated with Nr4a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a2
|
ENSMUSG00000026826.7 | nuclear receptor subfamily 4, group A, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a2 | mm10_v2_chr2_-_57113053_57113072 | -0.20 | 2.5e-01 | Click! |
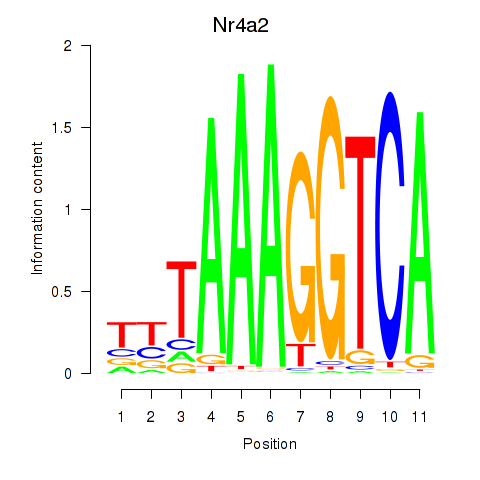
Activity profile of Nr4a2 motif
Sorted Z-values of Nr4a2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22002599 | 3.22 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr19_-_8405060 | 3.16 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr7_-_14438538 | 2.51 |
ENSMUST00000168252.2
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr1_+_167618246 | 2.13 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr10_-_92375367 | 2.09 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr8_+_104847061 | 1.94 |
ENSMUST00000055052.5
|
Ces2c
|
carboxylesterase 2C |
| chr6_-_59024340 | 1.59 |
ENSMUST00000173193.1
|
Fam13a
|
family with sequence similarity 13, member A |
| chr6_-_59024470 | 1.54 |
ENSMUST00000089860.5
|
Fam13a
|
family with sequence similarity 13, member A |
| chr8_-_64733534 | 1.46 |
ENSMUST00000141021.1
|
Sc4mol
|
sterol-C4-methyl oxidase-like |
| chr5_-_66151903 | 1.31 |
ENSMUST00000167950.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr13_-_92131494 | 1.26 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr10_-_42478280 | 1.26 |
ENSMUST00000151747.1
|
Lace1
|
lactation elevated 1 |
| chr5_+_30232581 | 1.17 |
ENSMUST00000145167.1
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr19_+_36554661 | 1.15 |
ENSMUST00000169036.2
ENSMUST00000047247.5 |
Hectd2
|
HECT domain containing 2 |
| chr5_-_66151323 | 1.14 |
ENSMUST00000131838.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr15_+_25940846 | 1.14 |
ENSMUST00000110438.1
|
Fam134b
|
family with sequence similarity 134, member B |
| chr10_-_42478488 | 1.10 |
ENSMUST00000041024.8
|
Lace1
|
lactation elevated 1 |
| chr4_-_42581621 | 1.03 |
ENSMUST00000178742.1
|
Gm10592
|
predicted gene 10592 |
| chr1_+_87574016 | 1.03 |
ENSMUST00000166259.1
ENSMUST00000172222.1 ENSMUST00000163606.1 |
Neu2
|
neuraminidase 2 |
| chr18_-_56975333 | 0.93 |
ENSMUST00000139243.2
ENSMUST00000025488.8 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr13_+_93303757 | 0.93 |
ENSMUST00000109494.1
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr4_-_141623799 | 0.92 |
ENSMUST00000038661.7
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr13_+_93304066 | 0.90 |
ENSMUST00000109493.1
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr5_+_102845007 | 0.89 |
ENSMUST00000070000.4
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr15_+_25773985 | 0.88 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr6_+_3993776 | 0.83 |
ENSMUST00000031673.5
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr9_-_22085391 | 0.79 |
ENSMUST00000179422.1
ENSMUST00000098937.3 ENSMUST00000177967.1 ENSMUST00000180180.1 |
Ecsit
|
ECSIT homolog (Drosophila) |
| chr2_-_73892588 | 0.78 |
ENSMUST00000154456.1
ENSMUST00000090802.4 ENSMUST00000055833.5 |
Atf2
|
activating transcription factor 2 |
| chr17_+_70522083 | 0.76 |
ENSMUST00000148486.1
ENSMUST00000133717.1 |
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr16_-_44139196 | 0.75 |
ENSMUST00000063661.6
ENSMUST00000114666.2 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr2_-_73892619 | 0.73 |
ENSMUST00000112007.1
ENSMUST00000112016.2 |
Atf2
|
activating transcription factor 2 |
| chr16_-_44139630 | 0.73 |
ENSMUST00000137557.1
ENSMUST00000147025.1 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr10_+_67185730 | 0.71 |
ENSMUST00000173689.1
|
Jmjd1c
|
jumonji domain containing 1C |
| chr13_+_74121435 | 0.70 |
ENSMUST00000036208.6
|
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr15_-_68363139 | 0.70 |
ENSMUST00000175699.1
|
Gm20732
|
predicted gene 20732 |
| chr2_+_61593077 | 0.69 |
ENSMUST00000112495.1
ENSMUST00000112501.2 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr7_-_141429351 | 0.68 |
ENSMUST00000164387.1
ENSMUST00000137488.1 ENSMUST00000084436.3 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr7_-_89338709 | 0.67 |
ENSMUST00000137723.1
ENSMUST00000117852.1 ENSMUST00000041968.3 |
Tmem135
|
transmembrane protein 135 |
| chrX_-_142390334 | 0.67 |
ENSMUST00000112907.1
|
Acsl4
|
acyl-CoA synthetase long-chain family member 4 |
| chr10_+_53337686 | 0.67 |
ENSMUST00000046221.6
ENSMUST00000163319.1 |
Pln
|
phospholamban |
| chr5_-_121527186 | 0.64 |
ENSMUST00000152270.1
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr19_+_3768112 | 0.63 |
ENSMUST00000005518.9
ENSMUST00000113967.1 ENSMUST00000152935.1 ENSMUST00000176262.1 ENSMUST00000176407.1 ENSMUST00000176926.1 ENSMUST00000176512.1 |
Suv420h1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr19_+_3767953 | 0.63 |
ENSMUST00000113970.1
|
Suv420h1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr1_-_124045247 | 0.61 |
ENSMUST00000112603.2
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr10_+_28074813 | 0.59 |
ENSMUST00000166468.1
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr2_+_30595037 | 0.59 |
ENSMUST00000102853.3
|
Cstad
|
CSA-conditional, T cell activation-dependent protein |
| chr12_+_31438209 | 0.59 |
ENSMUST00000001254.5
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr9_-_13446753 | 0.57 |
ENSMUST00000167906.2
|
Gm17571
|
predicted gene, 17571 |
| chr7_-_141429433 | 0.56 |
ENSMUST00000124444.1
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr3_+_122895072 | 0.56 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr1_-_24100306 | 0.55 |
ENSMUST00000027337.8
|
Fam135a
|
family with sequence similarity 135, member A |
| chr2_+_61593125 | 0.55 |
ENSMUST00000112494.1
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr4_-_150914401 | 0.54 |
ENSMUST00000105675.1
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr10_+_110920170 | 0.53 |
ENSMUST00000020403.5
|
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr3_-_144932523 | 0.51 |
ENSMUST00000098549.3
|
AI747448
|
expressed sequence AI747448 |
| chr16_+_44139821 | 0.50 |
ENSMUST00000159514.1
ENSMUST00000161326.1 ENSMUST00000063520.8 ENSMUST00000063542.7 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr7_+_105554360 | 0.50 |
ENSMUST00000046983.8
|
Smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr2_+_105668935 | 0.48 |
ENSMUST00000142772.1
|
Pax6
|
paired box gene 6 |
| chr17_-_35909626 | 0.46 |
ENSMUST00000141132.1
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr15_-_76229492 | 0.45 |
ENSMUST00000074834.5
|
Plec
|
plectin |
| chr17_-_43667015 | 0.45 |
ENSMUST00000024705.4
|
Slc25a27
|
solute carrier family 25, member 27 |
| chr4_+_102760294 | 0.45 |
ENSMUST00000072481.5
ENSMUST00000156596.1 ENSMUST00000080728.6 ENSMUST00000106882.2 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr8_-_13494479 | 0.44 |
ENSMUST00000033828.5
|
Gas6
|
growth arrest specific 6 |
| chr13_+_41655697 | 0.43 |
ENSMUST00000067176.8
|
Gm5082
|
predicted gene 5082 |
| chr2_-_120563795 | 0.43 |
ENSMUST00000055241.6
ENSMUST00000135625.1 |
Zfp106
|
zinc finger protein 106 |
| chr1_-_63176653 | 0.42 |
ENSMUST00000027111.8
ENSMUST00000168099.2 |
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr9_+_108392820 | 0.40 |
ENSMUST00000035234.4
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr11_-_100822525 | 0.40 |
ENSMUST00000107358.2
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr3_-_92500493 | 0.39 |
ENSMUST00000062129.1
|
Sprr4
|
small proline-rich protein 4 |
| chr2_+_4976113 | 0.39 |
ENSMUST00000167607.1
ENSMUST00000115010.2 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr2_+_4976176 | 0.38 |
ENSMUST00000027978.1
|
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr3_+_84925476 | 0.38 |
ENSMUST00000107675.1
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr12_-_76795489 | 0.37 |
ENSMUST00000082431.3
|
Gpx2
|
glutathione peroxidase 2 |
| chr19_+_43782181 | 0.36 |
ENSMUST00000026208.4
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr1_+_182409162 | 0.36 |
ENSMUST00000117245.1
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chr2_+_59160838 | 0.36 |
ENSMUST00000102754.4
ENSMUST00000168631.1 ENSMUST00000123908.1 |
Pkp4
|
plakophilin 4 |
| chr19_-_6921753 | 0.36 |
ENSMUST00000173635.1
|
Esrra
|
estrogen related receptor, alpha |
| chr10_+_94147982 | 0.34 |
ENSMUST00000105290.2
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr10_-_20725023 | 0.33 |
ENSMUST00000020165.7
|
Pde7b
|
phosphodiesterase 7B |
| chr10_-_41303171 | 0.33 |
ENSMUST00000043814.3
|
Fig4
|
FIG4 homolog (S. cerevisiae) |
| chr9_-_42461414 | 0.32 |
ENSMUST00000066179.7
|
Tbcel
|
tubulin folding cofactor E-like |
| chr6_+_113697050 | 0.31 |
ENSMUST00000089018.4
|
Tatdn2
|
TatD DNase domain containing 2 |
| chr3_+_14578609 | 0.31 |
ENSMUST00000029069.6
ENSMUST00000165922.2 |
E2f5
|
E2F transcription factor 5 |
| chrX_+_160768013 | 0.30 |
ENSMUST00000033650.7
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr5_+_63649335 | 0.30 |
ENSMUST00000159584.1
|
3110047P20Rik
|
RIKEN cDNA 3110047P20 gene |
| chr4_+_102760135 | 0.29 |
ENSMUST00000066824.7
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr15_+_102966794 | 0.29 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chr3_+_4211716 | 0.28 |
ENSMUST00000170943.1
|
Gm8775
|
predicted gene 8775 |
| chr6_-_124814288 | 0.28 |
ENSMUST00000172132.2
|
Tpi1
|
triosephosphate isomerase 1 |
| chr19_+_46152505 | 0.28 |
ENSMUST00000026254.7
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr7_+_4119556 | 0.28 |
ENSMUST00000079415.5
|
Ttyh1
|
tweety homolog 1 (Drosophila) |
| chr2_-_101621033 | 0.28 |
ENSMUST00000090513.4
|
B230118H07Rik
|
RIKEN cDNA B230118H07 gene |
| chr11_+_96282529 | 0.28 |
ENSMUST00000125410.1
|
Hoxb8
|
homeobox B8 |
| chr4_+_99955715 | 0.27 |
ENSMUST00000102783.4
|
Pgm2
|
phosphoglucomutase 2 |
| chr2_+_129065934 | 0.26 |
ENSMUST00000035812.7
|
Ttl
|
tubulin tyrosine ligase |
| chr10_+_127078886 | 0.26 |
ENSMUST00000039259.6
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr4_+_136357423 | 0.26 |
ENSMUST00000182167.1
|
Gm17388
|
predicted gene, 17388 |
| chr10_-_20724696 | 0.26 |
ENSMUST00000170265.1
|
Pde7b
|
phosphodiesterase 7B |
| chr7_+_120635176 | 0.26 |
ENSMUST00000033176.5
|
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr6_+_34920941 | 0.25 |
ENSMUST00000114999.1
|
Stra8
|
stimulated by retinoic acid gene 8 |
| chr1_-_191907527 | 0.24 |
ENSMUST00000069573.5
|
1700034H15Rik
|
RIKEN cDNA 1700034H15 gene |
| chr19_-_32061438 | 0.24 |
ENSMUST00000096119.4
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr2_+_144033059 | 0.24 |
ENSMUST00000037722.2
ENSMUST00000110032.1 |
Banf2
|
barrier to autointegration factor 2 |
| chr7_+_4119525 | 0.24 |
ENSMUST00000119661.1
ENSMUST00000129423.1 |
Ttyh1
|
tweety homolog 1 (Drosophila) |
| chr16_+_8830093 | 0.23 |
ENSMUST00000023150.5
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr2_-_73892530 | 0.23 |
ENSMUST00000136958.1
ENSMUST00000112010.2 ENSMUST00000128531.1 ENSMUST00000112017.1 |
Atf2
|
activating transcription factor 2 |
| chrX_-_48513518 | 0.22 |
ENSMUST00000114945.2
ENSMUST00000037349.7 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr8_+_70863127 | 0.21 |
ENSMUST00000050921.2
|
A230052G05Rik
|
RIKEN cDNA A230052G05 gene |
| chr3_-_88027465 | 0.21 |
ENSMUST00000005014.2
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr1_+_131910458 | 0.21 |
ENSMUST00000062264.6
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chrX_+_160768179 | 0.20 |
ENSMUST00000112368.2
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr17_-_35910032 | 0.19 |
ENSMUST00000141662.1
ENSMUST00000056034.6 ENSMUST00000077494.6 ENSMUST00000149277.1 ENSMUST00000061052.5 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr4_+_33081505 | 0.19 |
ENSMUST00000147889.1
|
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr2_-_129699833 | 0.19 |
ENSMUST00000028883.5
|
Pdyn
|
prodynorphin |
| chr2_+_79707780 | 0.19 |
ENSMUST00000090760.2
ENSMUST00000040863.4 ENSMUST00000111780.2 |
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chrX_+_7579666 | 0.18 |
ENSMUST00000115740.1
ENSMUST00000115739.1 |
Foxp3
|
forkhead box P3 |
| chr2_+_105668888 | 0.17 |
ENSMUST00000111086.4
ENSMUST00000111087.3 |
Pax6
|
paired box gene 6 |
| chr17_+_69439326 | 0.17 |
ENSMUST00000169935.1
|
A330050F15Rik
|
RIKEN cDNA A330050F15 gene |
| chr6_+_34920971 | 0.16 |
ENSMUST00000185102.1
ENSMUST00000114997.2 |
Stra8
|
stimulated by retinoic acid gene 8 |
| chr7_+_140920940 | 0.16 |
ENSMUST00000184560.1
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chrX_-_73921917 | 0.15 |
ENSMUST00000114389.3
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr9_+_118506226 | 0.15 |
ENSMUST00000084820.4
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr6_-_99096196 | 0.15 |
ENSMUST00000175886.1
|
Foxp1
|
forkhead box P1 |
| chr15_+_54952939 | 0.14 |
ENSMUST00000181704.1
|
Gm26684
|
predicted gene, 26684 |
| chr2_-_59160644 | 0.14 |
ENSMUST00000077687.5
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr17_-_17624458 | 0.13 |
ENSMUST00000041047.2
|
Lnpep
|
leucyl/cystinyl aminopeptidase |
| chr12_-_93929102 | 0.13 |
ENSMUST00000180321.1
|
Gm9726
|
predicted gene 9726 |
| chr3_+_89436736 | 0.12 |
ENSMUST00000146630.1
ENSMUST00000145753.1 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr17_-_25785533 | 0.11 |
ENSMUST00000140738.1
ENSMUST00000145053.1 ENSMUST00000138759.1 ENSMUST00000133071.1 ENSMUST00000077938.3 |
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chrX_-_73921930 | 0.10 |
ENSMUST00000033763.8
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr9_-_70934808 | 0.10 |
ENSMUST00000034731.8
|
Lipc
|
lipase, hepatic |
| chr17_-_25785324 | 0.10 |
ENSMUST00000150324.1
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chrX_-_73921828 | 0.09 |
ENSMUST00000096316.3
ENSMUST00000114390.1 ENSMUST00000114391.3 ENSMUST00000114387.1 |
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr13_+_94875600 | 0.09 |
ENSMUST00000022195.10
|
Otp
|
orthopedia homolog (Drosophila) |
| chr11_-_24075054 | 0.09 |
ENSMUST00000068360.1
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr2_-_26237368 | 0.09 |
ENSMUST00000036187.8
|
Qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr2_-_155945282 | 0.08 |
ENSMUST00000040162.2
|
Gdf5
|
growth differentiation factor 5 |
| chr1_-_87573825 | 0.08 |
ENSMUST00000068681.5
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr11_-_79504078 | 0.08 |
ENSMUST00000164465.2
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chrX_-_167855061 | 0.08 |
ENSMUST00000112146.1
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr10_+_90829835 | 0.08 |
ENSMUST00000179964.1
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr7_+_140920896 | 0.07 |
ENSMUST00000183845.1
ENSMUST00000106045.1 |
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr3_+_89436699 | 0.06 |
ENSMUST00000038942.3
ENSMUST00000130858.1 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr3_-_144975011 | 0.05 |
ENSMUST00000075496.4
ENSMUST00000029923.6 |
Clca6
|
chloride channel calcium activated 6 |
| chr7_-_133602110 | 0.05 |
ENSMUST00000033275.2
|
Tex36
|
testis expressed 36 |
| chr10_+_90829780 | 0.05 |
ENSMUST00000179337.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_+_57142782 | 0.04 |
ENSMUST00000139892.1
|
Megf10
|
multiple EGF-like-domains 10 |
| chr13_-_17805093 | 0.04 |
ENSMUST00000042365.7
|
Cdk13
|
cyclin-dependent kinase 13 |
| chr9_-_106656081 | 0.04 |
ENSMUST00000023959.7
|
Grm2
|
glutamate receptor, metabotropic 2 |
| chrX_+_152233228 | 0.03 |
ENSMUST00000112588.2
ENSMUST00000082177.6 |
Kdm5c
|
lysine (K)-specific demethylase 5C |
| chr10_+_93589413 | 0.03 |
ENSMUST00000181835.1
|
4933408J17Rik
|
RIKEN cDNA 4933408J17 gene |
| chr14_-_78088994 | 0.03 |
ENSMUST00000118785.2
ENSMUST00000066437.4 |
Fam216b
|
family with sequence similarity 216, member B |
| chr14_-_18893376 | 0.02 |
ENSMUST00000151926.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr12_+_87973528 | 0.01 |
ENSMUST00000110148.2
|
Gm2046
|
predicted gene 2046 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.3 | 1.7 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.3 | 1.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 2.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 3.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.9 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.2 | 3.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.7 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.7 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 1.0 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.5 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 1.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.1 | 0.9 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.3 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.2 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.7 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.4 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.6 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.2 | GO:0002465 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of histone deacetylation(GO:0031064) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.3 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 2.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.7 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.6 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.7 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.7 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.6 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.8 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.5 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 3.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 2.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 3.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.0 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 0.5 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.2 | 1.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 1.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.8 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |