Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
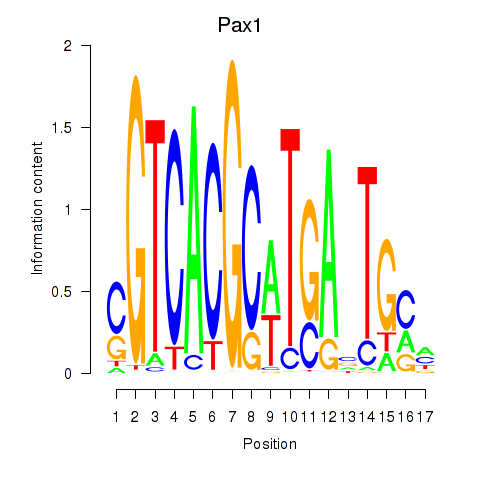
Results for Pax1_Pax9
Z-value: 0.38
Transcription factors associated with Pax1_Pax9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax1
|
ENSMUSG00000037034.9 | paired box 1 |
|
Pax9
|
ENSMUSG00000001497.12 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax9 | mm10_v2_chr12_+_56695628_56695700 | 0.44 | 7.4e-03 | Click! |
| Pax1 | mm10_v2_chr2_+_147364989_147365011 | 0.15 | 3.8e-01 | Click! |
Activity profile of Pax1_Pax9 motif
Sorted Z-values of Pax1_Pax9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_84734050 | 2.60 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr10_+_75573448 | 2.52 |
ENSMUST00000006508.3
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr9_-_22389113 | 1.57 |
ENSMUST00000040912.7
|
Anln
|
anillin, actin binding protein |
| chr5_+_112255813 | 1.54 |
ENSMUST00000031286.6
ENSMUST00000131673.1 ENSMUST00000112375.1 |
Crybb1
|
crystallin, beta B1 |
| chr9_+_96196246 | 1.15 |
ENSMUST00000165120.2
ENSMUST00000034982.9 |
Tfdp2
|
transcription factor Dp 2 |
| chr6_-_145048809 | 1.14 |
ENSMUST00000032402.5
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr13_-_62607499 | 0.95 |
ENSMUST00000091563.4
|
6720489N17Rik
|
RIKEN cDNA 6720489N17 gene |
| chr4_+_115000174 | 0.95 |
ENSMUST00000129957.1
|
Stil
|
Scl/Tal1 interrupting locus |
| chr4_+_115000156 | 0.90 |
ENSMUST00000030490.6
|
Stil
|
Scl/Tal1 interrupting locus |
| chr5_-_129787175 | 0.83 |
ENSMUST00000031399.6
|
Psph
|
phosphoserine phosphatase |
| chr1_-_37719782 | 0.81 |
ENSMUST00000160589.1
|
2010300C02Rik
|
RIKEN cDNA 2010300C02 gene |
| chr5_+_93206518 | 0.72 |
ENSMUST00000031330.4
|
2010109A12Rik
|
RIKEN cDNA 2010109A12 gene |
| chr9_-_97111117 | 0.65 |
ENSMUST00000085206.4
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr16_+_87553313 | 0.64 |
ENSMUST00000026700.7
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr7_-_24299310 | 0.51 |
ENSMUST00000145131.1
|
Zfp61
|
zinc finger protein 61 |
| chr13_-_62520451 | 0.41 |
ENSMUST00000082203.6
ENSMUST00000101547.4 |
Zfp934
|
zinc finger protein 934 |
| chr9_+_21337828 | 0.41 |
ENSMUST00000034697.7
|
Slc44a2
|
solute carrier family 44, member 2 |
| chr17_-_24205514 | 0.29 |
ENSMUST00000097376.3
ENSMUST00000168410.2 ENSMUST00000167791.2 ENSMUST00000040474.7 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr7_+_118633729 | 0.28 |
ENSMUST00000057320.7
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr4_+_11558914 | 0.28 |
ENSMUST00000178703.1
ENSMUST00000095145.5 ENSMUST00000108306.2 ENSMUST00000070755.6 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr13_-_62777089 | 0.26 |
ENSMUST00000167516.2
|
Gm5141
|
predicted gene 5141 |
| chr4_-_117375453 | 0.19 |
ENSMUST00000094853.2
|
Rnf220
|
ring finger protein 220 |
| chrX_+_74313014 | 0.15 |
ENSMUST00000114160.1
|
Fam50a
|
family with sequence similarity 50, member A |
| chr12_+_33957645 | 0.09 |
ENSMUST00000049089.5
|
Twist1
|
twist basic helix-loop-helix transcription factor 1 |
| chr8_+_31150307 | 0.09 |
ENSMUST00000098842.2
|
Tti2
|
TELO2 interacting protein 2 |
| chr4_-_143299463 | 0.04 |
ENSMUST00000119654.1
|
Pdpn
|
podoplanin |
| chr17_-_27909206 | 0.03 |
ENSMUST00000114848.1
|
Taf11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_+_129787390 | 0.01 |
ENSMUST00000031402.8
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chrX_+_86191764 | 0.01 |
ENSMUST00000026036.4
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax1_Pax9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 0.3 | 2.5 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.1 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 1.9 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.6 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 2.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |