Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
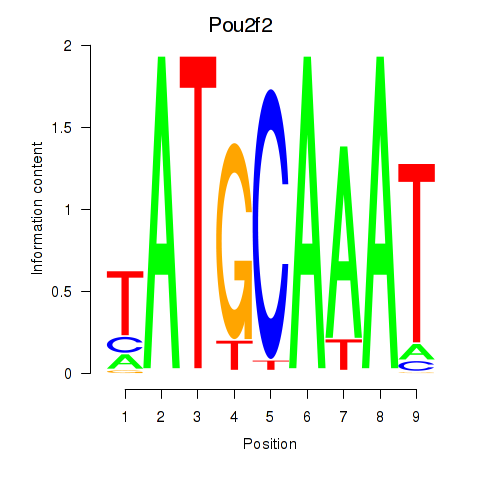
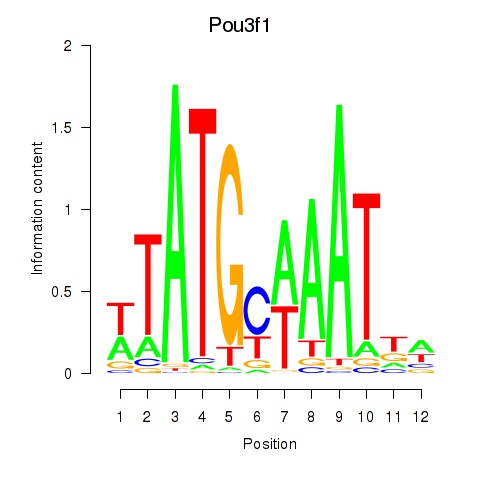
Results for Pou2f2_Pou3f1
Z-value: 1.42
Transcription factors associated with Pou2f2_Pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f2
|
ENSMUSG00000008496.12 | POU domain, class 2, transcription factor 2 |
|
Pou3f1
|
ENSMUSG00000090125.2 | POU domain, class 3, transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f1 | mm10_v2_chr4_+_124657646_124657656 | -0.29 | 9.1e-02 | Click! |
| Pou2f2 | mm10_v2_chr7_-_25132473_25132512 | 0.22 | 2.1e-01 | Click! |
Activity profile of Pou2f2_Pou3f1 motif
Sorted Z-values of Pou2f2_Pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23746734 | 11.16 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chr3_+_96269695 | 9.64 |
ENSMUST00000051089.3
ENSMUST00000177113.1 |
Hist2h2bb
|
histone cluster 2, H2bb |
| chr13_+_21722057 | 8.82 |
ENSMUST00000110476.3
|
Hist1h2bm
|
histone cluster 1, H2bm |
| chrX_+_93675088 | 6.71 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_+_58948890 | 6.42 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr10_-_80421847 | 5.57 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr7_-_102100227 | 5.55 |
ENSMUST00000106937.1
|
Art5
|
ADP-ribosyltransferase 5 |
| chr13_-_21833575 | 5.35 |
ENSMUST00000081342.5
|
Hist1h2ap
|
histone cluster 1, H2ap |
| chr16_+_17144600 | 5.07 |
ENSMUST00000115702.1
|
Ydjc
|
YdjC homolog (bacterial) |
| chr7_+_24507006 | 5.05 |
ENSMUST00000176880.1
|
Zfp428
|
zinc finger protein 428 |
| chr13_+_22043189 | 5.03 |
ENSMUST00000110452.1
|
Hist1h2bj
|
histone cluster 1, H2bj |
| chr13_+_23751069 | 4.89 |
ENSMUST00000078369.1
|
Hist1h2ab
|
histone cluster 1, H2ab |
| chr16_-_17144415 | 4.78 |
ENSMUST00000115709.1
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr5_+_76656512 | 4.76 |
ENSMUST00000086909.4
|
Gm10430
|
predicted gene 10430 |
| chr2_-_170194033 | 4.64 |
ENSMUST00000180625.1
|
Gm17619
|
predicted gene, 17619 |
| chr15_-_66831625 | 4.62 |
ENSMUST00000164163.1
|
Sla
|
src-like adaptor |
| chr7_+_24507122 | 4.60 |
ENSMUST00000177205.1
|
Zfp428
|
zinc finger protein 428 |
| chr6_-_67037399 | 4.59 |
ENSMUST00000043098.6
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr13_-_23571151 | 4.56 |
ENSMUST00000102969.3
|
Hist1h2ae
|
histone cluster 1, H2ae |
| chr13_-_22042949 | 4.52 |
ENSMUST00000091741.4
|
Hist1h2ag
|
histone cluster 1, H2ag |
| chr14_+_27000362 | 4.45 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr13_-_23683941 | 4.42 |
ENSMUST00000171127.1
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr11_+_95337012 | 4.00 |
ENSMUST00000037502.6
|
Fam117a
|
family with sequence similarity 117, member A |
| chr7_+_24507057 | 3.91 |
ENSMUST00000071361.6
|
Zfp428
|
zinc finger protein 428 |
| chr13_+_23544052 | 3.90 |
ENSMUST00000075558.2
|
Hist1h3f
|
histone cluster 1, H3f |
| chr3_+_51661167 | 3.79 |
ENSMUST00000099106.3
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr19_+_47228804 | 3.76 |
ENSMUST00000111807.3
|
Neurl1a
|
neuralized homolog 1A (Drosophila) |
| chr12_-_36042476 | 3.63 |
ENSMUST00000020896.8
|
Tspan13
|
tetraspanin 13 |
| chr11_+_58954675 | 3.56 |
ENSMUST00000108817.3
ENSMUST00000047697.5 |
Hist3h2a
Trim17
|
histone cluster 3, H2a tripartite motif-containing 17 |
| chr11_-_69605829 | 3.51 |
ENSMUST00000047889.6
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr8_+_22974844 | 3.50 |
ENSMUST00000110688.2
ENSMUST00000121802.2 |
Ank1
|
ankyrin 1, erythroid |
| chr1_+_91540553 | 3.45 |
ENSMUST00000027538.7
|
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr10_+_75948292 | 3.35 |
ENSMUST00000000926.2
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chrX_+_56454871 | 3.27 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr7_+_126776939 | 3.15 |
ENSMUST00000038614.5
ENSMUST00000170882.1 ENSMUST00000106359.1 ENSMUST00000106357.1 ENSMUST00000145762.1 ENSMUST00000132643.1 ENSMUST00000106356.1 |
Ypel3
|
yippee-like 3 (Drosophila) |
| chr1_-_89933290 | 3.10 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr13_+_22035821 | 3.00 |
ENSMUST00000110455.2
|
Hist1h2bk
|
histone cluster 1, H2bk |
| chr6_-_56901870 | 2.74 |
ENSMUST00000101367.2
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr12_-_114416895 | 2.68 |
ENSMUST00000179796.1
|
Ighv6-5
|
immunoglobulin heavy variable V6-5 |
| chr13_-_22035589 | 2.67 |
ENSMUST00000091742.4
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr13_+_23574381 | 2.64 |
ENSMUST00000090776.4
|
Hist1h2ad
|
histone cluster 1, H2ad |
| chr17_+_27556641 | 2.62 |
ENSMUST00000119486.1
ENSMUST00000118599.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr5_+_76840597 | 2.60 |
ENSMUST00000120639.2
ENSMUST00000163347.1 ENSMUST00000121851.1 |
C530008M17Rik
|
RIKEN cDNA C530008M17 gene |
| chr17_+_27556668 | 2.59 |
ENSMUST00000117254.1
ENSMUST00000118570.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr7_+_45017953 | 2.55 |
ENSMUST00000044111.7
|
Rras
|
Harvey rat sarcoma oncogene, subgroup R |
| chr7_+_24507099 | 2.50 |
ENSMUST00000177228.1
|
Zfp428
|
zinc finger protein 428 |
| chr17_+_27556613 | 2.47 |
ENSMUST00000117600.1
ENSMUST00000114888.3 |
Hmga1
|
high mobility group AT-hook 1 |
| chr9_+_21936986 | 2.45 |
ENSMUST00000046371.6
|
BC018242
|
cDNA sequence BC018242 |
| chr13_+_21810428 | 2.43 |
ENSMUST00000091745.5
|
Hist1h2ao
|
histone cluster 1, H2ao |
| chr4_-_128806045 | 2.43 |
ENSMUST00000106072.2
ENSMUST00000170934.1 |
Zfp362
|
zinc finger protein 362 |
| chrX_+_133850980 | 2.41 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr16_-_92697315 | 2.33 |
ENSMUST00000168195.1
ENSMUST00000113956.3 |
Runx1
|
runt related transcription factor 1 |
| chr7_-_102099932 | 2.33 |
ENSMUST00000106934.1
|
Art5
|
ADP-ribosyltransferase 5 |
| chr2_+_25180737 | 2.31 |
ENSMUST00000104999.2
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr15_+_62039216 | 2.26 |
ENSMUST00000183297.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr2_-_152398046 | 2.25 |
ENSMUST00000063332.8
ENSMUST00000182625.1 |
Sox12
|
SRY-box containing gene 12 |
| chr13_+_23533869 | 2.23 |
ENSMUST00000073261.2
|
Hist1h2af
|
histone cluster 1, H2af |
| chr11_-_106314494 | 2.21 |
ENSMUST00000167143.1
|
Cd79b
|
CD79B antigen |
| chr4_-_140810646 | 2.20 |
ENSMUST00000026377.2
|
Padi3
|
peptidyl arginine deiminase, type III |
| chr3_+_83766300 | 2.17 |
ENSMUST00000029625.7
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr7_+_127746775 | 2.15 |
ENSMUST00000033081.7
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr5_+_137629169 | 2.12 |
ENSMUST00000176667.1
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr4_+_152039315 | 2.07 |
ENSMUST00000084116.6
ENSMUST00000105663.1 ENSMUST00000103197.3 |
Nol9
|
nucleolar protein 9 |
| chr2_+_119047116 | 2.05 |
ENSMUST00000152380.1
ENSMUST00000099542.2 |
Casc5
|
cancer susceptibility candidate 5 |
| chr7_+_98838845 | 1.96 |
ENSMUST00000167303.1
|
Wnt11
|
wingless-related MMTV integration site 11 |
| chr16_-_92826004 | 1.91 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr1_-_144567667 | 1.91 |
ENSMUST00000184008.1
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr9_-_70421533 | 1.89 |
ENSMUST00000034742.6
|
Ccnb2
|
cyclin B2 |
| chrX_-_167209149 | 1.88 |
ENSMUST00000112176.1
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr3_-_50443603 | 1.85 |
ENSMUST00000029297.4
|
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr3_+_159495408 | 1.85 |
ENSMUST00000120272.1
ENSMUST00000029825.7 ENSMUST00000106041.2 |
Depdc1a
|
DEP domain containing 1a |
| chr13_+_5861489 | 1.84 |
ENSMUST00000000080.6
|
Klf6
|
Kruppel-like factor 6 |
| chr14_-_79771305 | 1.83 |
ENSMUST00000039568.5
|
Pcdh8
|
protocadherin 8 |
| chr5_+_137630116 | 1.82 |
ENSMUST00000175968.1
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr16_-_19883873 | 1.73 |
ENSMUST00000100083.3
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr12_-_79192248 | 1.71 |
ENSMUST00000161204.1
|
Rdh11
|
retinol dehydrogenase 11 |
| chr4_-_3938354 | 1.69 |
ENSMUST00000003369.3
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr9_+_71215779 | 1.68 |
ENSMUST00000034723.5
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr17_-_58991343 | 1.67 |
ENSMUST00000025064.7
|
2610034M16Rik
|
RIKEN cDNA 2610034M16 gene |
| chr10_+_79927039 | 1.66 |
ENSMUST00000019708.5
ENSMUST00000105377.1 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr19_+_8941865 | 1.65 |
ENSMUST00000096240.2
|
Mta2
|
metastasis-associated gene family, member 2 |
| chr10_+_79927330 | 1.65 |
ENSMUST00000105376.1
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr2_+_119047129 | 1.64 |
ENSMUST00000153300.1
ENSMUST00000028799.5 |
Casc5
|
cancer susceptibility candidate 5 |
| chr3_+_92365178 | 1.63 |
ENSMUST00000050397.1
|
Sprr2f
|
small proline-rich protein 2F |
| chr14_-_66868572 | 1.62 |
ENSMUST00000022629.8
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr3_-_144849301 | 1.60 |
ENSMUST00000159989.1
|
Clca4
|
chloride channel calcium activated 4 |
| chr6_+_136518820 | 1.59 |
ENSMUST00000032335.6
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_-_46044914 | 1.58 |
ENSMUST00000026252.7
|
Ldb1
|
LIM domain binding 1 |
| chr13_+_21833736 | 1.57 |
ENSMUST00000180288.1
ENSMUST00000110467.1 |
Hist1h2br
|
histone cluster 1 H2br |
| chr6_-_148946146 | 1.57 |
ENSMUST00000132696.1
|
Fam60a
|
family with sequence similarity 60, member A |
| chr6_-_52240841 | 1.56 |
ENSMUST00000121043.1
|
Hoxa10
|
homeobox A10 |
| chr3_+_130180882 | 1.54 |
ENSMUST00000106353.1
ENSMUST00000080335.4 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr5_-_147307264 | 1.53 |
ENSMUST00000031650.3
|
Cdx2
|
caudal type homeobox 2 |
| chr2_-_160912292 | 1.53 |
ENSMUST00000109454.1
ENSMUST00000057169.4 |
Emilin3
|
elastin microfibril interfacer 3 |
| chr7_-_143460989 | 1.52 |
ENSMUST00000167912.1
ENSMUST00000037287.6 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr13_-_21787218 | 1.52 |
ENSMUST00000091751.2
|
Hist1h2an
|
histone cluster 1, H2an |
| chr19_-_46045194 | 1.51 |
ENSMUST00000156585.1
ENSMUST00000152946.1 |
Ldb1
|
LIM domain binding 1 |
| chr12_-_114406773 | 1.49 |
ENSMUST00000177949.1
|
Ighv6-4
|
immunoglobulin heavy variable V6-4 |
| chr4_-_152477433 | 1.49 |
ENSMUST00000159186.1
ENSMUST00000162017.1 ENSMUST00000030768.2 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr8_+_120488416 | 1.46 |
ENSMUST00000034279.9
|
Gse1
|
genetic suppressor element 1 |
| chr6_-_13839916 | 1.46 |
ENSMUST00000060442.7
|
Gpr85
|
G protein-coupled receptor 85 |
| chr6_+_145145473 | 1.46 |
ENSMUST00000156849.1
ENSMUST00000132948.1 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr2_+_160645881 | 1.45 |
ENSMUST00000109468.2
|
Top1
|
topoisomerase (DNA) I |
| chr11_+_61208621 | 1.45 |
ENSMUST00000108716.1
ENSMUST00000019246.3 |
Aldh3a1
|
aldehyde dehydrogenase family 3, subfamily A1 |
| chr11_+_96282529 | 1.44 |
ENSMUST00000125410.1
|
Hoxb8
|
homeobox B8 |
| chr16_+_91729281 | 1.44 |
ENSMUST00000114001.1
ENSMUST00000113999.1 ENSMUST00000064797.5 ENSMUST00000114002.2 ENSMUST00000095909.3 ENSMUST00000056482.7 ENSMUST00000113996.1 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr8_-_122460666 | 1.38 |
ENSMUST00000006762.5
|
Snai3
|
snail homolog 3 (Drosophila) |
| chr3_+_92316705 | 1.36 |
ENSMUST00000061038.2
|
Sprr2b
|
small proline-rich protein 2B |
| chr9_-_22389113 | 1.35 |
ENSMUST00000040912.7
|
Anln
|
anillin, actin binding protein |
| chr17_+_29490812 | 1.34 |
ENSMUST00000024811.6
|
Pim1
|
proviral integration site 1 |
| chr12_-_17324703 | 1.33 |
ENSMUST00000020884.9
ENSMUST00000095820.5 ENSMUST00000127185.1 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr7_+_82174796 | 1.31 |
ENSMUST00000032874.7
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr11_-_34833631 | 1.30 |
ENSMUST00000093191.2
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr2_-_152830266 | 1.29 |
ENSMUST00000140436.1
|
Bcl2l1
|
BCL2-like 1 |
| chr5_-_123865491 | 1.26 |
ENSMUST00000057145.5
|
Niacr1
|
niacin receptor 1 |
| chr6_+_129350237 | 1.25 |
ENSMUST00000065289.4
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr4_+_129960760 | 1.25 |
ENSMUST00000139884.1
|
1700003M07Rik
|
RIKEN cDNA 1700003M07 gene |
| chr5_-_65091584 | 1.25 |
ENSMUST00000043352.4
|
Tmem156
|
transmembrane protein 156 |
| chr2_-_152830615 | 1.24 |
ENSMUST00000146380.1
ENSMUST00000134902.1 ENSMUST00000134357.1 ENSMUST00000109820.3 |
Bcl2l1
|
BCL2-like 1 |
| chr11_+_96282648 | 1.23 |
ENSMUST00000168043.1
|
Hoxb8
|
homeobox B8 |
| chr2_-_102451792 | 1.21 |
ENSMUST00000099678.3
|
Fjx1
|
four jointed box 1 (Drosophila) |
| chr8_-_70234401 | 1.15 |
ENSMUST00000019679.5
|
Armc6
|
armadillo repeat containing 6 |
| chr11_-_96747405 | 1.13 |
ENSMUST00000180492.1
|
2010300F17Rik
|
RIKEN cDNA 2010300F17 gene |
| chr1_+_52008210 | 1.12 |
ENSMUST00000027277.5
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr7_+_25282179 | 1.11 |
ENSMUST00000163320.1
ENSMUST00000005578.6 |
Cic
|
capicua homolog (Drosophila) |
| chr11_+_98741805 | 1.10 |
ENSMUST00000064187.5
|
Thra
|
thyroid hormone receptor alpha |
| chr13_-_21810190 | 1.08 |
ENSMUST00000110469.1
ENSMUST00000091749.2 |
Hist1h2bq
|
histone cluster 1, H2bq |
| chr17_-_35164891 | 1.07 |
ENSMUST00000025253.5
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr2_+_91096744 | 1.06 |
ENSMUST00000132741.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr15_+_85017138 | 1.06 |
ENSMUST00000023070.5
|
Upk3a
|
uroplakin 3A |
| chr7_-_102477902 | 1.04 |
ENSMUST00000061482.5
|
Olfr543
|
olfactory receptor 543 |
| chr9_+_74848437 | 1.01 |
ENSMUST00000161862.1
ENSMUST00000162089.1 ENSMUST00000160017.1 ENSMUST00000160950.1 |
Gm16551
Gm20649
|
predicted gene 16551 predicted gene 20649 |
| chr6_+_122553799 | 1.00 |
ENSMUST00000043301.7
|
Aicda
|
activation-induced cytidine deaminase |
| chr17_-_46556158 | 0.97 |
ENSMUST00000015749.5
|
Srf
|
serum response factor |
| chr7_+_27486910 | 0.96 |
ENSMUST00000008528.7
|
Sertad1
|
SERTA domain containing 1 |
| chr18_+_53551594 | 0.96 |
ENSMUST00000115398.1
|
Prdm6
|
PR domain containing 6 |
| chr12_-_114060315 | 0.96 |
ENSMUST00000103469.2
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr6_-_65144908 | 0.95 |
ENSMUST00000031982.4
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr6_+_30568367 | 0.94 |
ENSMUST00000049251.5
|
Cpa4
|
carboxypeptidase A4 |
| chr13_-_49309217 | 0.92 |
ENSMUST00000110087.2
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chrX_-_164250368 | 0.90 |
ENSMUST00000112263.1
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr8_+_70234613 | 0.89 |
ENSMUST00000145078.1
|
Sugp2
|
SURP and G patch domain containing 2 |
| chr14_-_67715585 | 0.87 |
ENSMUST00000163100.1
ENSMUST00000132705.1 ENSMUST00000124045.1 |
Cdca2
|
cell division cycle associated 2 |
| chr11_+_98741871 | 0.86 |
ENSMUST00000103139.4
|
Thra
|
thyroid hormone receptor alpha |
| chr16_+_17233560 | 0.86 |
ENSMUST00000090190.5
ENSMUST00000115698.2 |
Hic2
|
hypermethylated in cancer 2 |
| chr1_+_12718496 | 0.86 |
ENSMUST00000088585.3
|
Sulf1
|
sulfatase 1 |
| chr11_+_60469339 | 0.84 |
ENSMUST00000071880.2
ENSMUST00000081823.5 ENSMUST00000094135.2 |
Myo15
|
myosin XV |
| chr17_-_27133902 | 0.83 |
ENSMUST00000119227.1
ENSMUST00000025045.8 |
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr4_-_137118135 | 0.80 |
ENSMUST00000154285.1
|
Gm13001
|
predicted gene 13001 |
| chr6_-_142804390 | 0.80 |
ENSMUST00000111768.1
|
Gm766
|
predicted gene 766 |
| chr4_-_88033328 | 0.79 |
ENSMUST00000078090.5
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr8_+_31150307 | 0.77 |
ENSMUST00000098842.2
|
Tti2
|
TELO2 interacting protein 2 |
| chr1_-_126830632 | 0.75 |
ENSMUST00000112583.1
ENSMUST00000094609.3 |
Nckap5
|
NCK-associated protein 5 |
| chr11_+_51289920 | 0.74 |
ENSMUST00000102765.2
|
Col23a1
|
collagen, type XXIII, alpha 1 |
| chr11_-_5837760 | 0.73 |
ENSMUST00000109837.1
|
Polm
|
polymerase (DNA directed), mu |
| chr2_-_71367749 | 0.71 |
ENSMUST00000151937.1
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr8_-_122678653 | 0.68 |
ENSMUST00000134045.1
|
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) |
| chr6_-_56369625 | 0.67 |
ENSMUST00000170774.1
ENSMUST00000168944.1 ENSMUST00000166890.1 |
Pde1c
|
phosphodiesterase 1C |
| chr7_+_141195047 | 0.67 |
ENSMUST00000047093.4
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr16_-_4880284 | 0.67 |
ENSMUST00000037843.6
|
Ubald1
|
UBA-like domain containing 1 |
| chr1_+_75236439 | 0.67 |
ENSMUST00000082158.6
ENSMUST00000055223.7 |
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr18_+_50051702 | 0.66 |
ENSMUST00000134348.1
ENSMUST00000153873.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr17_+_47140942 | 0.66 |
ENSMUST00000077951.7
|
Trerf1
|
transcriptional regulating factor 1 |
| chr7_+_122289297 | 0.66 |
ENSMUST00000064989.5
ENSMUST00000064921.4 |
Prkcb
|
protein kinase C, beta |
| chr11_-_98053415 | 0.66 |
ENSMUST00000017544.2
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr2_+_74681991 | 0.65 |
ENSMUST00000142312.1
|
Hoxd11
|
homeobox D11 |
| chr8_-_9771018 | 0.65 |
ENSMUST00000110969.3
|
Fam155a
|
family with sequence similarity 155, member A |
| chr19_+_41593363 | 0.64 |
ENSMUST00000099454.3
|
AI606181
|
expressed sequence AI606181 |
| chr4_+_123183722 | 0.63 |
ENSMUST00000152194.1
|
Hpcal4
|
hippocalcin-like 4 |
| chr19_+_53140430 | 0.62 |
ENSMUST00000111741.2
|
Add3
|
adducin 3 (gamma) |
| chr12_-_115964196 | 0.61 |
ENSMUST00000103550.2
|
Ighv1-83
|
immunoglobulin heavy variable 1-83 |
| chr17_-_24533709 | 0.61 |
ENSMUST00000061764.7
|
Rab26
|
RAB26, member RAS oncogene family |
| chr7_+_82175156 | 0.58 |
ENSMUST00000180243.1
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr7_-_28372494 | 0.57 |
ENSMUST00000119990.1
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr5_-_122988533 | 0.57 |
ENSMUST00000086200.4
ENSMUST00000156474.1 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr17_+_27057288 | 0.57 |
ENSMUST00000049308.8
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr14_+_62292475 | 0.55 |
ENSMUST00000166879.1
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr4_-_64046925 | 0.55 |
ENSMUST00000107377.3
|
Tnc
|
tenascin C |
| chr14_+_79515618 | 0.54 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chrX_-_111531163 | 0.54 |
ENSMUST00000128819.1
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr13_+_21787461 | 0.53 |
ENSMUST00000110473.2
ENSMUST00000102982.1 |
Hist1h2bp
|
histone cluster 1, H2bp |
| chr1_+_70725902 | 0.53 |
ENSMUST00000161937.1
ENSMUST00000162182.1 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr4_+_128654686 | 0.51 |
ENSMUST00000030588.6
ENSMUST00000136377.1 |
Phc2
|
polyhomeotic-like 2 (Drosophila) |
| chr13_+_83738874 | 0.51 |
ENSMUST00000052354.4
|
C130071C03Rik
|
RIKEN cDNA C130071C03 gene |
| chr18_-_80986578 | 0.50 |
ENSMUST00000057950.7
|
Sall3
|
sal-like 3 (Drosophila) |
| chr10_+_90071095 | 0.49 |
ENSMUST00000183109.1
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr19_-_40994133 | 0.48 |
ENSMUST00000117695.1
|
Blnk
|
B cell linker |
| chr12_-_115790884 | 0.48 |
ENSMUST00000081809.5
|
Ighv1-73
|
immunoglobulin heavy variable 1-73 |
| chr16_-_89818338 | 0.46 |
ENSMUST00000164263.2
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr1_+_70725715 | 0.46 |
ENSMUST00000053922.5
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr5_-_122989086 | 0.46 |
ENSMUST00000046073.9
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_+_53590215 | 0.46 |
ENSMUST00000037618.6
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_-_122900267 | 0.46 |
ENSMUST00000031435.7
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chrX_+_150594420 | 0.45 |
ENSMUST00000112713.2
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr7_+_27591513 | 0.44 |
ENSMUST00000108344.2
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr16_-_91728599 | 0.42 |
ENSMUST00000122254.1
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr13_-_23621090 | 0.41 |
ENSMUST00000091704.5
ENSMUST00000051091.3 |
Hist1h2be
|
histone cluster 1, H2be |
| chr4_+_104766308 | 0.41 |
ENSMUST00000031663.3
|
C8b
|
complement component 8, beta polypeptide |
| chr9_-_85327110 | 0.41 |
ENSMUST00000034802.8
|
Fam46a
|
family with sequence similarity 46, member A |
| chr3_+_76074270 | 0.40 |
ENSMUST00000038364.8
|
Fstl5
|
follistatin-like 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f2_Pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.9 | 3.5 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.8 | 3.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.7 | 2.2 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.7 | 5.6 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.7 | 2.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.7 | 2.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.6 | 2.5 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.6 | 4.6 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.5 | 2.0 | GO:0060775 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.5 | 3.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.4 | 1.3 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 0.4 | 4.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.4 | 2.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 1.7 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 4.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 44.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 0.7 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 | 3.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 3.4 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.3 | 1.0 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 3.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 0.9 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.3 | 3.9 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 1.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 1.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.2 | 1.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.2 | 25.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 1.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 2.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 1.3 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.2 | 0.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 1.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 1.9 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 0.4 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.2 | 2.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 7.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 1.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 1.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 0.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 1.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 2.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 7.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.2 | 0.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.2 | 3.8 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 0.5 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.2 | 6.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.3 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 2.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 3.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.6 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 2.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 1.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 1.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 5.5 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 0.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 1.6 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 1.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 3.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.8 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.3 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.1 | 4.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 0.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.3 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 2.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.2 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 1.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.0 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.5 | GO:0021891 | olfactory bulb interneuron differentiation(GO:0021889) olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 1.6 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 1.4 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.9 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.4 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 2.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.7 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 1.5 | GO:0007338 | single fertilization(GO:0007338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.6 | 47.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 3.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 8.4 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.3 | 1.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 1.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 2.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 21.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.0 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 1.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 3.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 6.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.1 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 1.3 | 6.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.9 | 3.8 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.8 | 7.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.7 | 5.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.7 | 2.0 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.4 | 2.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 1.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.4 | 3.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 3.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 2.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.3 | 1.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 3.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 6.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 2.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 0.6 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 1.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 0.6 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.6 | GO:0005220 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.7 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 5.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 4.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.8 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 3.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.2 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 9.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 2.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 21.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.5 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 9.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 4.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 2.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 3.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 43.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 1.0 | 7.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 6.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 5.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 2.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 3.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 4.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 3.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |