Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pou3f3
Z-value: 0.80
Transcription factors associated with Pou3f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f3
|
ENSMUSG00000045515.2 | POU domain, class 3, transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f3 | mm10_v2_chr1_+_42697146_42697146 | 0.27 | 1.1e-01 | Click! |
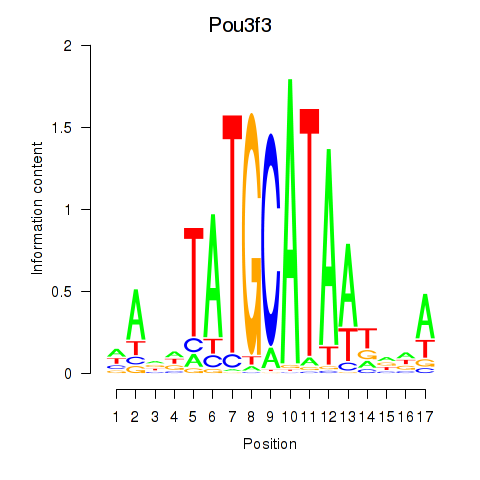
Activity profile of Pou3f3 motif
Sorted Z-values of Pou3f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_76656512 | 9.48 |
ENSMUST00000086909.4
|
Gm10430
|
predicted gene 10430 |
| chr14_+_27000362 | 6.60 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr5_+_66676098 | 6.28 |
ENSMUST00000031131.9
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr4_+_134510999 | 5.98 |
ENSMUST00000105866.2
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr12_-_114416895 | 5.04 |
ENSMUST00000179796.1
|
Ighv6-5
|
immunoglobulin heavy variable V6-5 |
| chr4_-_149137536 | 4.27 |
ENSMUST00000176124.1
ENSMUST00000177408.1 ENSMUST00000105695.1 ENSMUST00000030813.3 |
Apitd1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr3_+_83766300 | 4.11 |
ENSMUST00000029625.7
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr11_+_9191934 | 3.73 |
ENSMUST00000042740.6
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr3_-_123034943 | 2.79 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr6_+_129350237 | 2.49 |
ENSMUST00000065289.4
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr17_-_50094277 | 1.99 |
ENSMUST00000113195.1
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr15_-_42676967 | 1.90 |
ENSMUST00000022921.5
|
Angpt1
|
angiopoietin 1 |
| chr2_+_25180737 | 1.83 |
ENSMUST00000104999.2
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr11_-_99244058 | 1.39 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr5_-_74065736 | 1.14 |
ENSMUST00000145016.1
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr7_-_126792469 | 0.73 |
ENSMUST00000032936.6
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr3_-_144849301 | 0.51 |
ENSMUST00000159989.1
|
Clca4
|
chloride channel calcium activated 4 |
| chr2_+_110721587 | 0.47 |
ENSMUST00000111017.2
|
Muc15
|
mucin 15 |
| chr3_-_146108047 | 0.47 |
ENSMUST00000160285.1
|
Wdr63
|
WD repeat domain 63 |
| chrX_+_112011007 | 0.44 |
ENSMUST00000131304.1
|
Tex16
|
testis expressed gene 16 |
| chr10_+_26229707 | 0.36 |
ENSMUST00000060716.5
ENSMUST00000164660.1 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr8_-_105295934 | 0.35 |
ENSMUST00000057855.3
|
Exoc3l
|
exocyst complex component 3-like |
| chr5_-_66514815 | 0.06 |
ENSMUST00000161879.1
ENSMUST00000159357.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.0 | 6.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.7 | 6.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.6 | 1.9 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 2.0 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.8 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 5.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 6.0 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 2.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.7 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 6.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 6.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 2.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 4.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 5.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 6.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 3.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 2.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 4.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |