Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Prdm1
Z-value: 1.38
Transcription factors associated with Prdm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm1
|
ENSMUSG00000038151.6 | PR domain containing 1, with ZNF domain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm1 | mm10_v2_chr10_-_44458715_44458751 | -0.56 | 4.1e-04 | Click! |
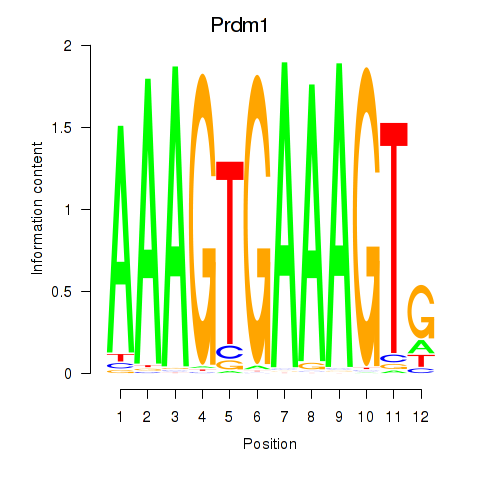
Activity profile of Prdm1 motif
Sorted Z-values of Prdm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_77729091 | 11.02 |
ENSMUST00000109775.2
|
Apol9b
|
apolipoprotein L 9b |
| chr8_+_70083509 | 8.71 |
ENSMUST00000007738.9
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr6_-_55175019 | 7.78 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr11_-_5950018 | 7.42 |
ENSMUST00000102920.3
|
Gck
|
glucokinase |
| chr5_+_30921825 | 7.08 |
ENSMUST00000117435.1
|
Khk
|
ketohexokinase |
| chr5_+_30921867 | 6.77 |
ENSMUST00000123885.1
|
Khk
|
ketohexokinase |
| chr5_+_30921556 | 6.67 |
ENSMUST00000031053.8
|
Khk
|
ketohexokinase |
| chr4_-_42756543 | 6.27 |
ENSMUST00000102957.3
|
Ccl19
|
chemokine (C-C motif) ligand 19 |
| chr1_-_139608282 | 5.89 |
ENSMUST00000170441.2
|
Cfhr3
|
complement factor H-related 3 |
| chr19_-_11050500 | 5.39 |
ENSMUST00000099676.4
|
AW112010
|
expressed sequence AW112010 |
| chr4_+_42629719 | 5.37 |
ENSMUST00000166898.2
|
Gm2564
|
predicted gene 2564 |
| chr16_+_43363855 | 4.55 |
ENSMUST00000156367.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr19_+_44203265 | 4.33 |
ENSMUST00000026220.5
|
Scd3
|
stearoyl-coenzyme A desaturase 3 |
| chr7_-_29248375 | 4.07 |
ENSMUST00000032808.4
|
2200002D01Rik
|
RIKEN cDNA 2200002D01 gene |
| chr2_-_126783416 | 3.93 |
ENSMUST00000130356.1
ENSMUST00000028842.2 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr5_-_105239533 | 3.77 |
ENSMUST00000065588.6
|
Gbp10
|
guanylate-binding protein 10 |
| chr3_-_113574758 | 3.73 |
ENSMUST00000106540.1
|
Amy1
|
amylase 1, salivary |
| chr9_-_106476372 | 3.57 |
ENSMUST00000123555.1
ENSMUST00000125850.1 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr10_-_109009055 | 3.53 |
ENSMUST00000156979.1
|
Syt1
|
synaptotagmin I |
| chr11_+_75468040 | 3.39 |
ENSMUST00000043598.7
ENSMUST00000108435.1 |
Tlcd2
|
TLC domain containing 2 |
| chr3_+_122895072 | 3.30 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr9_-_106476590 | 3.29 |
ENSMUST00000112479.2
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr16_+_43364145 | 3.29 |
ENSMUST00000148775.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_-_106476104 | 3.18 |
ENSMUST00000156426.1
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr14_+_41105359 | 3.15 |
ENSMUST00000047286.6
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr8_-_84800344 | 3.12 |
ENSMUST00000099070.3
|
Nfix
|
nuclear factor I/X |
| chr4_-_96591555 | 3.03 |
ENSMUST00000055693.8
|
Cyp2j9
|
cytochrome P450, family 2, subfamily j, polypeptide 9 |
| chr11_-_73324616 | 3.02 |
ENSMUST00000021119.2
|
Aspa
|
aspartoacylase |
| chr8_-_84800024 | 3.01 |
ENSMUST00000126806.1
ENSMUST00000076715.6 |
Nfix
|
nuclear factor I/X |
| chr16_-_24393588 | 2.99 |
ENSMUST00000181640.1
|
1110054M08Rik
|
RIKEN cDNA 1110054M08 gene |
| chr17_+_37193889 | 2.98 |
ENSMUST00000038844.6
|
Ubd
|
ubiquitin D |
| chr4_-_108071327 | 2.95 |
ENSMUST00000106701.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr15_-_89379246 | 2.93 |
ENSMUST00000049968.7
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr5_+_114923234 | 2.85 |
ENSMUST00000031540.4
ENSMUST00000112143.3 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr4_-_148152059 | 2.79 |
ENSMUST00000056965.5
ENSMUST00000168503.1 ENSMUST00000152098.1 |
Fbxo6
|
F-box protein 6 |
| chr15_+_25843264 | 2.76 |
ENSMUST00000022881.7
|
Fam134b
|
family with sequence similarity 134, member B |
| chr2_+_43555321 | 2.75 |
ENSMUST00000028223.2
|
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr10_-_95324072 | 2.74 |
ENSMUST00000053594.5
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr10_+_116301374 | 2.73 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr9_-_107668967 | 2.70 |
ENSMUST00000177567.1
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr15_+_66891477 | 2.69 |
ENSMUST00000118823.1
|
Wisp1
|
WNT1 inducible signaling pathway protein 1 |
| chr8_+_94525067 | 2.69 |
ENSMUST00000098489.4
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr10_-_75797528 | 2.54 |
ENSMUST00000120177.1
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr3_-_101604580 | 2.53 |
ENSMUST00000036493.6
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr1_-_106759727 | 2.53 |
ENSMUST00000010049.4
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr2_+_116067213 | 2.52 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr19_-_11081088 | 2.50 |
ENSMUST00000025636.6
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr14_-_26066961 | 2.49 |
ENSMUST00000100818.5
|
Tmem254c
|
transmembrane protein 254c |
| chr3_-_108044801 | 2.48 |
ENSMUST00000178808.1
ENSMUST00000106670.1 ENSMUST00000029489.8 |
Gstm4
|
glutathione S-transferase, mu 4 |
| chr15_+_66891320 | 2.45 |
ENSMUST00000005255.2
|
Wisp1
|
WNT1 inducible signaling pathway protein 1 |
| chr2_+_43555342 | 2.45 |
ENSMUST00000112826.1
ENSMUST00000050511.6 |
Kynu
|
kynureninase (L-kynurenine hydrolase) |
| chr19_+_34640871 | 2.40 |
ENSMUST00000102824.3
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr11_+_103103051 | 2.39 |
ENSMUST00000152971.1
|
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr4_+_42255767 | 2.38 |
ENSMUST00000178864.1
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr9_-_44802951 | 2.34 |
ENSMUST00000044694.6
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr13_+_33004528 | 2.32 |
ENSMUST00000006391.4
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr13_-_12464925 | 2.28 |
ENSMUST00000124888.1
|
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr13_+_74639866 | 2.28 |
ENSMUST00000169114.1
|
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr13_+_23839401 | 2.24 |
ENSMUST00000039721.7
ENSMUST00000166467.1 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr6_+_117168535 | 2.22 |
ENSMUST00000112866.1
ENSMUST00000112871.1 ENSMUST00000073043.4 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr14_-_25927250 | 2.22 |
ENSMUST00000100811.5
|
Tmem254a
|
transmembrane protein 254a |
| chr7_-_141435327 | 2.20 |
ENSMUST00000138865.1
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr11_-_53859187 | 2.18 |
ENSMUST00000117316.1
ENSMUST00000120776.1 ENSMUST00000121435.1 |
Gm12216
|
predicted gene 12216 |
| chr4_-_42773993 | 2.18 |
ENSMUST00000095114.4
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
| chr1_-_172590463 | 2.16 |
ENSMUST00000065679.6
|
Slamf8
|
SLAM family member 8 |
| chr3_+_145758674 | 2.16 |
ENSMUST00000029845.8
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr10_-_75797728 | 2.14 |
ENSMUST00000139724.1
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr8_+_127064022 | 2.11 |
ENSMUST00000160272.1
ENSMUST00000079777.5 ENSMUST00000162907.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr2_+_69135799 | 2.05 |
ENSMUST00000041865.7
|
Nostrin
|
nitric oxide synthase trafficker |
| chr10_-_78352323 | 2.03 |
ENSMUST00000001240.5
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr15_+_32920723 | 2.03 |
ENSMUST00000022871.5
|
Sdc2
|
syndecan 2 |
| chr10_-_78351711 | 2.03 |
ENSMUST00000105390.1
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr8_+_127064107 | 1.98 |
ENSMUST00000162536.1
ENSMUST00000026921.6 ENSMUST00000162665.1 ENSMUST00000160766.1 ENSMUST00000162602.1 ENSMUST00000162531.1 ENSMUST00000160581.1 ENSMUST00000161355.1 ENSMUST00000159537.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr14_-_16249154 | 1.98 |
ENSMUST00000148121.1
ENSMUST00000112624.1 |
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr11_-_77894096 | 1.98 |
ENSMUST00000017597.4
|
Pipox
|
pipecolic acid oxidase |
| chr5_+_92137896 | 1.96 |
ENSMUST00000031355.6
|
Uso1
|
USO1 vesicle docking factor |
| chr1_-_184845838 | 1.94 |
ENSMUST00000068725.3
|
Marc2
|
mitochondrial amidoxime reducing component 2 |
| chr3_+_81932601 | 1.85 |
ENSMUST00000029649.2
|
Ctso
|
cathepsin O |
| chr11_+_70459940 | 1.84 |
ENSMUST00000147289.1
ENSMUST00000126105.1 |
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr16_+_97536079 | 1.84 |
ENSMUST00000024112.7
|
Mx2
|
myxovirus (influenza virus) resistance 2 |
| chr9_-_45204083 | 1.84 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr18_-_12879973 | 1.84 |
ENSMUST00000119512.1
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr17_-_12916345 | 1.82 |
ENSMUST00000079121.3
|
Mrpl18
|
mitochondrial ribosomal protein L18 |
| chr17_-_34187219 | 1.79 |
ENSMUST00000173831.1
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr15_+_44787746 | 1.78 |
ENSMUST00000181839.1
|
2310069G16Rik
|
RIKEN cDNA 2310069G16 gene |
| chr15_+_99392948 | 1.75 |
ENSMUST00000161250.1
ENSMUST00000160635.1 ENSMUST00000161778.1 |
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr8_+_22283441 | 1.73 |
ENSMUST00000077194.1
|
Tpte
|
transmembrane phosphatase with tensin homology |
| chr18_+_84851338 | 1.72 |
ENSMUST00000160180.1
|
Cyb5
|
cytochrome b-5 |
| chr7_-_5413145 | 1.69 |
ENSMUST00000108569.2
|
Vmn1r58
|
vomeronasal 1 receptor 58 |
| chr9_-_78322357 | 1.67 |
ENSMUST00000095071.4
|
Gm8074
|
predicted gene 8074 |
| chrX_+_161717055 | 1.67 |
ENSMUST00000112338.1
|
Rai2
|
retinoic acid induced 2 |
| chr17_+_33919332 | 1.66 |
ENSMUST00000025161.7
|
Tapbp
|
TAP binding protein |
| chr1_-_140183283 | 1.66 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr10_+_81574699 | 1.65 |
ENSMUST00000131794.1
ENSMUST00000136341.1 |
Tle2
|
transducin-like enhancer of split 2, homolog of Drosophila E(spl) |
| chr16_-_35871544 | 1.65 |
ENSMUST00000042665.8
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr17_+_84956718 | 1.64 |
ENSMUST00000112305.3
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr2_-_101621033 | 1.64 |
ENSMUST00000090513.4
|
B230118H07Rik
|
RIKEN cDNA B230118H07 gene |
| chr10_-_78352053 | 1.63 |
ENSMUST00000105388.1
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr9_-_98955302 | 1.62 |
ENSMUST00000181706.1
|
Foxl2os
|
forkhead box L2 opposite strand transcript |
| chr1_+_153899937 | 1.60 |
ENSMUST00000086199.5
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr16_+_24393350 | 1.58 |
ENSMUST00000038053.6
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr5_+_63649335 | 1.56 |
ENSMUST00000159584.1
|
3110047P20Rik
|
RIKEN cDNA 3110047P20 gene |
| chr7_+_30184160 | 1.56 |
ENSMUST00000098594.2
|
Cox7a1
|
cytochrome c oxidase subunit VIIa 1 |
| chr9_-_106891401 | 1.55 |
ENSMUST00000069036.7
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr16_+_11008898 | 1.54 |
ENSMUST00000180624.1
|
Gm4262
|
predicted gene 4262 |
| chr14_-_45477856 | 1.52 |
ENSMUST00000141424.1
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr2_+_97467657 | 1.52 |
ENSMUST00000059049.7
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr8_-_94434259 | 1.52 |
ENSMUST00000180445.1
|
9330175E14Rik
|
RIKEN cDNA 9330175E14 gene |
| chr14_-_101640434 | 1.51 |
ENSMUST00000168587.1
|
Commd6
|
COMM domain containing 6 |
| chr5_-_93206489 | 1.48 |
ENSMUST00000058550.8
|
Ccni
|
cyclin I |
| chr6_+_114648811 | 1.47 |
ENSMUST00000182510.1
|
Atg7
|
autophagy related 7 |
| chr8_+_88697022 | 1.45 |
ENSMUST00000043526.8
|
Cyld
|
cylindromatosis (turban tumor syndrome) |
| chr2_+_76675265 | 1.45 |
ENSMUST00000111920.1
|
Plekha3
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 3 |
| chr14_+_16249259 | 1.44 |
ENSMUST00000022310.6
|
Ngly1
|
N-glycanase 1 |
| chr17_-_25785533 | 1.39 |
ENSMUST00000140738.1
ENSMUST00000145053.1 ENSMUST00000138759.1 ENSMUST00000133071.1 ENSMUST00000077938.3 |
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr11_+_120677226 | 1.39 |
ENSMUST00000129644.2
ENSMUST00000151160.1 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr16_-_30267524 | 1.38 |
ENSMUST00000064856.7
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr9_-_106891965 | 1.37 |
ENSMUST00000159283.1
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr4_-_84546284 | 1.37 |
ENSMUST00000177040.1
|
Bnc2
|
basonuclin 2 |
| chr12_+_37241633 | 1.36 |
ENSMUST00000049874.7
|
Agmo
|
alkylglycerol monooxygenase |
| chr14_-_101640670 | 1.35 |
ENSMUST00000100339.2
|
Commd6
|
COMM domain containing 6 |
| chr11_+_49076584 | 1.34 |
ENSMUST00000109202.1
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr10_+_4710119 | 1.33 |
ENSMUST00000105588.1
ENSMUST00000105589.1 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr17_-_35895920 | 1.32 |
ENSMUST00000059740.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr10_+_128322443 | 1.30 |
ENSMUST00000026446.2
|
Cnpy2
|
canopy 2 homolog (zebrafish) |
| chr8_-_94434142 | 1.29 |
ENSMUST00000180612.1
|
9330175E14Rik
|
RIKEN cDNA 9330175E14 gene |
| chr10_-_78351690 | 1.29 |
ENSMUST00000166360.1
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr9_-_29963112 | 1.28 |
ENSMUST00000075069.4
|
Ntm
|
neurotrimin |
| chr1_-_184033998 | 1.27 |
ENSMUST00000050306.5
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr3_-_86548268 | 1.25 |
ENSMUST00000077524.3
|
Mab21l2
|
mab-21-like 2 (C. elegans) |
| chr1_+_139501692 | 1.25 |
ENSMUST00000027615.5
|
F13b
|
coagulation factor XIII, beta subunit |
| chr3_-_88425094 | 1.25 |
ENSMUST00000168755.1
ENSMUST00000057935.6 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr4_-_45489794 | 1.24 |
ENSMUST00000146236.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr12_-_72664759 | 1.24 |
ENSMUST00000021512.9
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr4_-_63745055 | 1.24 |
ENSMUST00000062246.6
|
Tnfsf15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr15_-_76243401 | 1.22 |
ENSMUST00000165738.1
ENSMUST00000075689.6 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr5_-_135962275 | 1.21 |
ENSMUST00000054895.3
|
Srcrb4d
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr2_+_164332740 | 1.21 |
ENSMUST00000017148.7
|
Svs5
|
seminal vesicle secretory protein 5 |
| chr7_+_131371138 | 1.20 |
ENSMUST00000075610.6
|
Pstk
|
phosphoseryl-tRNA kinase |
| chr17_-_32034493 | 1.19 |
ENSMUST00000002145.5
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr3_+_142701067 | 1.19 |
ENSMUST00000044392.4
|
Ccbl2
|
cysteine conjugate-beta lyase 2 |
| chr8_-_45410539 | 1.18 |
ENSMUST00000034056.4
ENSMUST00000167106.1 |
Tlr3
|
toll-like receptor 3 |
| chr9_+_38718263 | 1.17 |
ENSMUST00000001544.5
ENSMUST00000118144.1 |
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr3_-_7613427 | 1.17 |
ENSMUST00000168269.2
|
Il7
|
interleukin 7 |
| chr7_-_27166413 | 1.16 |
ENSMUST00000108382.1
|
Egln2
|
EGL nine homolog 2 (C. elegans) |
| chr10_-_78352212 | 1.12 |
ENSMUST00000146899.1
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr3_-_65392579 | 1.11 |
ENSMUST00000029414.5
|
Ssr3
|
signal sequence receptor, gamma |
| chr13_-_18031616 | 1.10 |
ENSMUST00000099736.2
|
Vdac3-ps1
|
voltage-dependent anion channel 3, pseudogene 1 |
| chr4_-_148151646 | 1.09 |
ENSMUST00000132083.1
|
Fbxo6
|
F-box protein 6 |
| chrX_-_74023908 | 1.08 |
ENSMUST00000033769.8
ENSMUST00000114352.1 ENSMUST00000068286.5 ENSMUST00000114360.3 ENSMUST00000114354.3 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr1_-_78196832 | 1.08 |
ENSMUST00000004994.9
|
Pax3
|
paired box gene 3 |
| chr6_-_39118211 | 1.07 |
ENSMUST00000038398.6
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr8_-_93363676 | 1.06 |
ENSMUST00000145041.1
|
Ces1h
|
carboxylesterase 1H |
| chr4_-_52497244 | 1.06 |
ENSMUST00000114578.4
|
Vma21-ps
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae), pseudogene |
| chr7_+_100706623 | 1.05 |
ENSMUST00000107042.1
|
Fam168a
|
family with sequence similarity 168, member A |
| chr15_-_103340085 | 1.03 |
ENSMUST00000168828.1
|
Zfp385a
|
zinc finger protein 385A |
| chr13_-_92483996 | 1.02 |
ENSMUST00000040106.7
|
Fam151b
|
family with sequence similarity 151, member B |
| chr4_+_141115660 | 1.02 |
ENSMUST00000181450.1
|
4921514A10Rik
|
RIKEN cDNA 4921514A10 gene |
| chr17_+_24895116 | 1.02 |
ENSMUST00000043907.7
|
Mrps34
|
mitochondrial ribosomal protein S34 |
| chr8_-_129065488 | 1.01 |
ENSMUST00000125112.1
ENSMUST00000108747.2 ENSMUST00000095158.4 |
Ccdc7
|
coiled-coil domain containing 7 |
| chr3_+_142701044 | 1.01 |
ENSMUST00000106218.1
|
Ccbl2
|
cysteine conjugate-beta lyase 2 |
| chr18_-_66022580 | 1.01 |
ENSMUST00000143990.1
|
Lman1
|
lectin, mannose-binding, 1 |
| chr3_+_106482427 | 0.99 |
ENSMUST00000029508.4
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr2_-_51973219 | 0.99 |
ENSMUST00000028314.2
|
Nmi
|
N-myc (and STAT) interactor |
| chr11_+_72301613 | 0.98 |
ENSMUST00000151440.1
ENSMUST00000146233.1 ENSMUST00000140842.2 |
Xaf1
|
XIAP associated factor 1 |
| chr8_+_46739745 | 0.97 |
ENSMUST00000034041.7
|
Irf2
|
interferon regulatory factor 2 |
| chr1_-_78197112 | 0.96 |
ENSMUST00000087086.6
|
Pax3
|
paired box gene 3 |
| chr7_-_45136102 | 0.96 |
ENSMUST00000125500.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr1_+_184034381 | 0.95 |
ENSMUST00000048655.7
|
Dusp10
|
dual specificity phosphatase 10 |
| chr10_-_75517856 | 0.94 |
ENSMUST00000123505.1
ENSMUST00000147269.1 ENSMUST00000118936.1 |
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr13_+_30974204 | 0.93 |
ENSMUST00000070258.2
|
Gm5447
|
predicted gene 5447 |
| chr1_+_181150926 | 0.92 |
ENSMUST00000134115.1
ENSMUST00000111059.1 |
Cnih4
|
cornichon homolog 4 (Drosophila) |
| chr4_-_72200833 | 0.92 |
ENSMUST00000102848.2
ENSMUST00000072695.6 ENSMUST00000107337.1 ENSMUST00000074216.7 |
Tle1
|
transducin-like enhancer of split 1, homolog of Drosophila E(spl) |
| chr12_+_8208107 | 0.92 |
ENSMUST00000037383.5
ENSMUST00000169104.1 |
1110057K04Rik
|
RIKEN cDNA 1110057K04 gene |
| chr2_-_90904827 | 0.92 |
ENSMUST00000005647.3
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr10_-_116950366 | 0.91 |
ENSMUST00000020375.6
|
Rab3ip
|
RAB3A interacting protein |
| chr3_-_98564038 | 0.91 |
ENSMUST00000058728.5
|
Gm10681
|
predicted gene 10681 |
| chr14_+_64652524 | 0.90 |
ENSMUST00000100473.4
|
Kif13b
|
kinesin family member 13B |
| chr1_+_6730135 | 0.89 |
ENSMUST00000155921.1
|
St18
|
suppression of tumorigenicity 18 |
| chr14_+_122534305 | 0.89 |
ENSMUST00000154206.1
ENSMUST00000038374.6 ENSMUST00000135578.1 |
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide |
| chr5_+_28165690 | 0.89 |
ENSMUST00000036177.7
|
En2
|
engrailed 2 |
| chr1_+_132008285 | 0.88 |
ENSMUST00000146432.1
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr1_-_67038824 | 0.86 |
ENSMUST00000119559.1
ENSMUST00000149996.1 ENSMUST00000027149.5 ENSMUST00000113979.3 |
Lancl1
|
LanC (bacterial lantibiotic synthetase component C)-like 1 |
| chr6_+_71373400 | 0.85 |
ENSMUST00000066747.7
ENSMUST00000172321.1 |
Cd8a
|
CD8 antigen, alpha chain |
| chr6_-_125231772 | 0.85 |
ENSMUST00000043422.7
|
Tapbpl
|
TAP binding protein-like |
| chr5_-_93206428 | 0.85 |
ENSMUST00000144514.1
|
Ccni
|
cyclin I |
| chr17_-_66519666 | 0.85 |
ENSMUST00000167962.1
ENSMUST00000070538.4 |
Rab12
|
RAB12, member RAS oncogene family |
| chr12_+_37242030 | 0.84 |
ENSMUST00000160390.1
|
Agmo
|
alkylglycerol monooxygenase |
| chr4_+_150236816 | 0.84 |
ENSMUST00000080926.6
|
Eno1
|
enolase 1, alpha non-neuron |
| chr7_-_45136391 | 0.84 |
ENSMUST00000146760.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr7_-_45136056 | 0.83 |
ENSMUST00000130628.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr6_-_129451906 | 0.82 |
ENSMUST00000037481.7
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr1_+_180942500 | 0.82 |
ENSMUST00000159436.1
|
Tmem63a
|
transmembrane protein 63a |
| chr6_+_147042755 | 0.82 |
ENSMUST00000036111.7
|
Mrps35
|
mitochondrial ribosomal protein S35 |
| chr12_+_37241729 | 0.81 |
ENSMUST00000160768.1
|
Agmo
|
alkylglycerol monooxygenase |
| chr1_+_6730051 | 0.80 |
ENSMUST00000043578.6
ENSMUST00000131467.1 ENSMUST00000150761.1 ENSMUST00000151281.1 |
St18
|
suppression of tumorigenicity 18 |
| chr1_-_130887408 | 0.79 |
ENSMUST00000121040.1
|
Il24
|
interleukin 24 |
| chr11_-_70459957 | 0.79 |
ENSMUST00000019064.2
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr8_-_83332416 | 0.79 |
ENSMUST00000177594.1
ENSMUST00000053902.3 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr17_-_24895047 | 0.78 |
ENSMUST00000119848.1
ENSMUST00000121542.1 |
Eme2
|
essential meiotic endonuclease 1 homolog 2 (S. pombe) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.5 | GO:0009744 | fructose catabolic process(GO:0006001) response to sucrose(GO:0009744) response to disaccharide(GO:0034285) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 3.3 | 10.0 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 2.7 | 8.2 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.7 | 5.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.6 | 4.7 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 1.5 | 7.4 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 1.1 | 4.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.0 | 2.9 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.9 | 2.7 | GO:2000485 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) regulation of glutamine transport(GO:2000485) |
| 0.9 | 0.9 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.8 | 3.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.7 | 3.0 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.7 | 2.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.7 | 4.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.7 | 2.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.6 | 2.5 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.6 | 1.8 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.6 | 2.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.6 | 1.7 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.6 | 2.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.5 | 1.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.5 | 3.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.5 | 2.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.5 | 1.5 | GO:0039521 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.4 | 3.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.4 | 1.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.4 | 1.3 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.4 | 2.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 1.5 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.3 | 1.7 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 1.0 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.3 | 1.0 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.3 | 6.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 2.7 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.3 | 3.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 3.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 2.5 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.3 | 0.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.3 | 2.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 1.6 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.3 | 5.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.3 | 1.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 2.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.2 | 0.7 | GO:0000966 | RNA 5'-end processing(GO:0000966) mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 1.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 1.9 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.2 | 1.2 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.2 | 5.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.2 | 2.2 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.2 | 3.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 2.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 3.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 1.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 0.7 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 7.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 11.8 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.2 | 0.7 | GO:0070268 | cornification(GO:0070268) |
| 0.2 | 0.5 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 | 2.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.7 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.2 | 0.8 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.2 | 0.9 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.2 | 2.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 1.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 2.5 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 0.6 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.5 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.1 | 1.2 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.9 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.1 | 0.5 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.5 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.4 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.4 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 3.3 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.3 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.3 | GO:2000836 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) positive regulation of androgen secretion(GO:2000836) |
| 0.1 | 1.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 2.6 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 0.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 0.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.4 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 1.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 7.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 1.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 8.1 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.7 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.9 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.8 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 2.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.8 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 1.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.8 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0042697 | menopause(GO:0042697) |
| 0.0 | 0.3 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.5 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 2.0 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 1.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 7.4 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.9 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.7 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 7.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 2.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 3.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.3 | 2.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 9.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 3.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 3.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 0.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 2.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 1.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 28.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.0 | 6.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.5 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 1.6 | 6.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.6 | 4.7 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 1.3 | 5.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.3 | 7.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.1 | 10.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.1 | 4.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.0 | 3.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.0 | 3.0 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.9 | 2.7 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.9 | 3.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.7 | 3.0 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.7 | 7.4 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.6 | 1.9 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.5 | 3.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.5 | 1.6 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.5 | 2.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.5 | 2.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 1.5 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.5 | 3.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 1.3 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 8.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 13.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.3 | 8.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 2.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 1.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 0.9 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.3 | 1.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 2.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 2.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 2.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 4.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 1.7 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.2 | 1.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 5.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 3.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 2.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.8 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 2.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 1.5 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 2.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 5.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 5.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 2.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 3.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 3.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 4.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 2.5 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.3 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 8.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 3.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 7.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 5.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 7.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 3.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 7.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.2 | 9.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 8.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 4.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 1.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 23.6 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 4.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 2.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |