Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
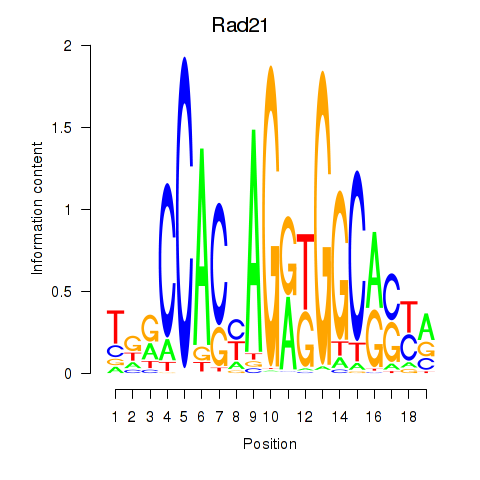
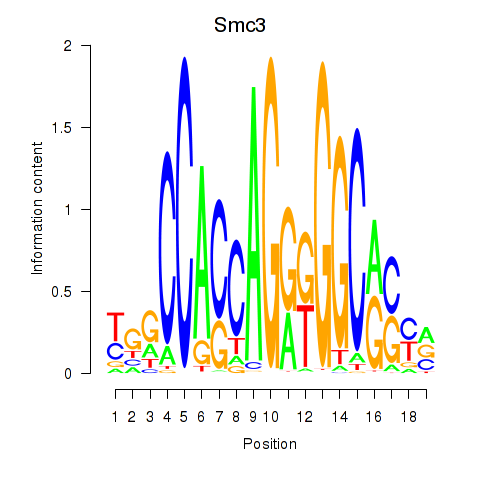
Results for Rad21_Smc3
Z-value: 1.18
Transcription factors associated with Rad21_Smc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rad21
|
ENSMUSG00000022314.9 | RAD21 cohesin complex component |
|
Smc3
|
ENSMUSG00000024974.10 | structural maintenance of chromosomes 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rad21 | mm10_v2_chr15_-_51991679_51991760 | -0.87 | 6.6e-12 | Click! |
| Smc3 | mm10_v2_chr19_+_53600377_53600435 | -0.79 | 1.1e-08 | Click! |
Activity profile of Rad21_Smc3 motif
Sorted Z-values of Rad21_Smc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_45069137 | 9.58 |
ENSMUST00000067984.7
|
Mtnr1a
|
melatonin receptor 1A |
| chr15_+_10314102 | 8.31 |
ENSMUST00000127467.1
|
Prlr
|
prolactin receptor |
| chr7_-_19698383 | 8.29 |
ENSMUST00000173739.1
|
Apoe
|
apolipoprotein E |
| chr7_-_19698206 | 8.23 |
ENSMUST00000172808.1
ENSMUST00000174191.1 |
Apoe
|
apolipoprotein E |
| chr18_-_61911783 | 7.48 |
ENSMUST00000049378.8
ENSMUST00000166783.1 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr11_+_72435565 | 7.28 |
ENSMUST00000100903.2
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr5_+_30921556 | 6.92 |
ENSMUST00000031053.8
|
Khk
|
ketohexokinase |
| chr19_+_42036025 | 6.76 |
ENSMUST00000026172.2
|
Ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr6_+_124570294 | 6.75 |
ENSMUST00000184647.1
|
C1rb
|
complement component 1, r subcomponent B |
| chr6_+_124512615 | 6.57 |
ENSMUST00000068593.7
|
C1ra
|
complement component 1, r subcomponent A |
| chr5_+_30921825 | 6.55 |
ENSMUST00000117435.1
|
Khk
|
ketohexokinase |
| chr1_+_182564994 | 6.20 |
ENSMUST00000048941.7
ENSMUST00000168514.1 |
Capn8
|
calpain 8 |
| chr8_+_45069374 | 6.02 |
ENSMUST00000130141.1
|
Mtnr1a
|
melatonin receptor 1A |
| chr9_+_111439063 | 5.96 |
ENSMUST00000111879.3
|
Dclk3
|
doublecortin-like kinase 3 |
| chr5_+_30921867 | 5.77 |
ENSMUST00000123885.1
|
Khk
|
ketohexokinase |
| chr2_+_31887262 | 5.67 |
ENSMUST00000138325.1
ENSMUST00000028187.6 |
Lamc3
|
laminin gamma 3 |
| chr6_-_85915653 | 5.58 |
ENSMUST00000161198.2
|
Cml1
|
camello-like 1 |
| chr4_+_134396320 | 5.24 |
ENSMUST00000105869.2
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr6_-_85915604 | 5.22 |
ENSMUST00000174369.1
|
Cml1
|
camello-like 1 |
| chr10_-_31445921 | 5.01 |
ENSMUST00000000305.5
|
Tpd52l1
|
tumor protein D52-like 1 |
| chr5_-_151369172 | 4.51 |
ENSMUST00000067770.3
|
D730045B01Rik
|
RIKEN cDNA D730045B01 gene |
| chr11_+_103103051 | 4.48 |
ENSMUST00000152971.1
|
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr3_+_135281221 | 4.32 |
ENSMUST00000120397.1
ENSMUST00000171974.1 ENSMUST00000029817.7 |
Bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr1_-_51915968 | 4.09 |
ENSMUST00000046390.7
|
Myo1b
|
myosin IB |
| chr7_+_127800604 | 4.05 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr7_+_127800844 | 4.01 |
ENSMUST00000106271.1
ENSMUST00000138432.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr5_-_130024280 | 3.94 |
ENSMUST00000161640.1
ENSMUST00000161884.1 ENSMUST00000161094.1 |
Asl
|
argininosuccinate lyase |
| chr18_-_3281036 | 3.86 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr4_-_57143437 | 3.77 |
ENSMUST00000095076.3
ENSMUST00000030142.3 |
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b |
| chr4_-_149307506 | 3.74 |
ENSMUST00000055647.8
ENSMUST00000030806.5 ENSMUST00000060537.6 |
Kif1b
|
kinesin family member 1B |
| chr8_+_60506109 | 3.65 |
ENSMUST00000079472.2
|
Aadat
|
aminoadipate aminotransferase |
| chr7_-_34655500 | 3.64 |
ENSMUST00000032709.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr6_+_85915787 | 3.63 |
ENSMUST00000149026.1
|
Tprkb
|
Tp53rk binding protein |
| chr5_+_151368683 | 3.52 |
ENSMUST00000181114.1
ENSMUST00000181555.1 |
1700028E10Rik
|
RIKEN cDNA 1700028E10 gene |
| chr9_+_100643605 | 3.43 |
ENSMUST00000041418.6
|
Stag1
|
stromal antigen 1 |
| chr12_-_102704896 | 3.42 |
ENSMUST00000178697.1
ENSMUST00000046518.5 |
Itpk1
|
inositol 1,3,4-triphosphate 5/6 kinase |
| chr6_+_85915709 | 3.41 |
ENSMUST00000113751.1
ENSMUST00000113753.1 ENSMUST00000113752.1 ENSMUST00000067137.7 |
Tprkb
|
Tp53rk binding protein |
| chr7_+_64501687 | 3.37 |
ENSMUST00000032732.8
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr2_-_166155272 | 3.32 |
ENSMUST00000088086.3
|
Sulf2
|
sulfatase 2 |
| chr4_-_114908892 | 3.27 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr2_+_152669461 | 3.26 |
ENSMUST00000125366.1
ENSMUST00000109825.1 ENSMUST00000089059.2 ENSMUST00000079247.3 |
H13
|
histocompatibility 13 |
| chr2_+_25428699 | 3.21 |
ENSMUST00000102919.3
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr4_+_100095791 | 3.13 |
ENSMUST00000039630.5
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr10_+_29313164 | 3.12 |
ENSMUST00000160399.1
|
Echdc1
|
enoyl Coenzyme A hydratase domain containing 1 |
| chr2_-_173119402 | 3.11 |
ENSMUST00000094287.3
ENSMUST00000179693.1 |
Ctcfl
|
CCCTC-binding factor (zinc finger protein)-like |
| chr7_+_64502090 | 2.97 |
ENSMUST00000137732.1
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr14_+_65666394 | 2.94 |
ENSMUST00000022610.8
|
Scara5
|
scavenger receptor class A, member 5 (putative) |
| chr3_+_118562129 | 2.93 |
ENSMUST00000039177.7
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr4_+_41762309 | 2.91 |
ENSMUST00000108042.2
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr10_+_29313500 | 2.91 |
ENSMUST00000020034.4
|
Echdc1
|
enoyl Coenzyme A hydratase domain containing 1 |
| chr10_+_29313227 | 2.87 |
ENSMUST00000161605.1
|
Echdc1
|
enoyl Coenzyme A hydratase domain containing 1 |
| chr19_-_28963863 | 2.86 |
ENSMUST00000161813.1
|
4430402I18Rik
|
RIKEN cDNA 4430402I18 gene |
| chr2_+_58567360 | 2.84 |
ENSMUST00000071543.5
|
Upp2
|
uridine phosphorylase 2 |
| chr7_+_64501949 | 2.84 |
ENSMUST00000138829.1
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr7_+_35119285 | 2.75 |
ENSMUST00000042985.9
|
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr3_-_63929376 | 2.74 |
ENSMUST00000061706.6
|
E130311K13Rik
|
RIKEN cDNA E130311K13 gene |
| chr16_+_18052860 | 2.74 |
ENSMUST00000143343.1
|
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr7_-_127935429 | 2.73 |
ENSMUST00000141385.1
ENSMUST00000156152.1 |
Prss36
|
protease, serine, 36 |
| chr17_+_34204080 | 2.68 |
ENSMUST00000138491.1
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr7_-_19749464 | 2.67 |
ENSMUST00000075447.7
ENSMUST00000108450.3 |
Pvrl2
|
poliovirus receptor-related 2 |
| chr5_-_25100624 | 2.65 |
ENSMUST00000030784.7
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_-_44790146 | 2.60 |
ENSMUST00000115056.1
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr13_+_51100810 | 2.56 |
ENSMUST00000095797.5
|
Spin1
|
spindlin 1 |
| chr19_+_5878622 | 2.55 |
ENSMUST00000136833.1
ENSMUST00000141362.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr13_+_64248649 | 2.54 |
ENSMUST00000181403.1
|
1810034E14Rik
|
RIKEN cDNA 1810034E14 gene |
| chr9_+_100643755 | 2.53 |
ENSMUST00000133388.1
|
Stag1
|
stromal antigen 1 |
| chr9_-_107710475 | 2.52 |
ENSMUST00000080560.3
|
Sema3f
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr4_-_141618238 | 2.52 |
ENSMUST00000053263.8
|
Tmem82
|
transmembrane protein 82 |
| chr18_+_12643329 | 2.50 |
ENSMUST00000025294.7
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr8_+_119446719 | 2.49 |
ENSMUST00000098363.3
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr13_-_35027077 | 2.48 |
ENSMUST00000170538.1
ENSMUST00000163280.1 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr7_+_16309577 | 2.48 |
ENSMUST00000002152.6
|
Bbc3
|
BCL2 binding component 3 |
| chr4_+_122995944 | 2.47 |
ENSMUST00000106252.2
|
Mycl
|
v-myc myelocytomatosis viral oncogene homolog, lung carcinoma derived (avian) |
| chrX_-_44790179 | 2.46 |
ENSMUST00000060481.2
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr8_-_69890967 | 2.45 |
ENSMUST00000152938.1
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr9_+_100643448 | 2.45 |
ENSMUST00000146312.1
ENSMUST00000129269.1 |
Stag1
|
stromal antigen 1 |
| chr3_+_54735536 | 2.45 |
ENSMUST00000044567.3
|
Alg5
|
asparagine-linked glycosylation 5 (dolichyl-phosphate beta-glucosyltransferase) |
| chr2_+_70661556 | 2.45 |
ENSMUST00000112201.1
ENSMUST00000028509.4 ENSMUST00000133432.1 ENSMUST00000112205.1 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr9_+_45906513 | 2.45 |
ENSMUST00000039059.6
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr8_+_22192800 | 2.45 |
ENSMUST00000033866.8
|
Vps36
|
vacuolar protein sorting 36 (yeast) |
| chr10_-_128409632 | 2.38 |
ENSMUST00000172348.1
ENSMUST00000166608.1 ENSMUST00000164199.1 ENSMUST00000171370.1 ENSMUST00000026439.7 |
Nabp2
|
nucleic acid binding protein 2 |
| chr7_+_44896125 | 2.37 |
ENSMUST00000166552.1
ENSMUST00000168207.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chr19_+_5877794 | 2.37 |
ENSMUST00000145200.1
ENSMUST00000025732.7 ENSMUST00000125114.1 ENSMUST00000155697.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr4_+_41755210 | 2.27 |
ENSMUST00000108038.1
ENSMUST00000084695.4 |
Galt
|
galactose-1-phosphate uridyl transferase |
| chrX_-_140600497 | 2.27 |
ENSMUST00000112996.2
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr15_-_76232554 | 2.21 |
ENSMUST00000166428.1
|
Plec
|
plectin |
| chr14_-_70520254 | 2.18 |
ENSMUST00000022693.7
|
Bmp1
|
bone morphogenetic protein 1 |
| chr7_+_16310412 | 2.18 |
ENSMUST00000136781.1
|
Bbc3
|
BCL2 binding component 3 |
| chr1_+_16105774 | 2.17 |
ENSMUST00000027053.7
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr14_+_65666430 | 2.15 |
ENSMUST00000069226.6
|
Scara5
|
scavenger receptor class A, member 5 (putative) |
| chr10_-_10558199 | 2.15 |
ENSMUST00000019974.3
|
Rab32
|
RAB32, member RAS oncogene family |
| chr18_-_3337467 | 2.14 |
ENSMUST00000154135.1
|
Crem
|
cAMP responsive element modulator |
| chr11_-_109298121 | 2.14 |
ENSMUST00000020920.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr15_-_77399086 | 2.13 |
ENSMUST00000175919.1
ENSMUST00000176074.1 |
Apol7a
|
apolipoprotein L 7a |
| chr19_+_5406815 | 2.13 |
ENSMUST00000174412.1
ENSMUST00000153017.2 |
4930481A15Rik
|
RIKEN cDNA 4930481A15 gene |
| chr15_+_99099412 | 2.12 |
ENSMUST00000061295.6
|
Dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr1_+_191575721 | 2.06 |
ENSMUST00000045450.5
|
Ints7
|
integrator complex subunit 7 |
| chr19_+_6418731 | 2.04 |
ENSMUST00000113462.1
ENSMUST00000077182.6 ENSMUST00000113461.1 |
Nrxn2
|
neurexin II |
| chr14_+_77036746 | 2.01 |
ENSMUST00000048208.3
ENSMUST00000095625.4 |
Ccdc122
|
coiled-coil domain containing 122 |
| chr18_-_3337614 | 1.99 |
ENSMUST00000150235.1
ENSMUST00000154470.1 |
Crem
|
cAMP responsive element modulator |
| chr14_+_52810934 | 1.98 |
ENSMUST00000103646.3
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr2_-_65567505 | 1.96 |
ENSMUST00000100069.2
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr9_-_31211805 | 1.93 |
ENSMUST00000072634.7
ENSMUST00000079758.7 |
Aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr11_+_104231390 | 1.92 |
ENSMUST00000106992.3
|
Mapt
|
microtubule-associated protein tau |
| chr8_+_111536492 | 1.92 |
ENSMUST00000168428.1
ENSMUST00000171182.1 |
Znrf1
|
zinc and ring finger 1 |
| chr18_-_37935378 | 1.92 |
ENSMUST00000025337.7
|
Diap1
|
diaphanous homolog 1 (Drosophila) |
| chr6_-_129237948 | 1.91 |
ENSMUST00000181238.1
ENSMUST00000180379.1 |
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene |
| chr10_-_75773350 | 1.89 |
ENSMUST00000001716.7
|
Ddt
|
D-dopachrome tautomerase |
| chr19_-_47464406 | 1.89 |
ENSMUST00000111800.2
ENSMUST00000081619.2 |
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr18_-_37935403 | 1.89 |
ENSMUST00000080033.6
ENSMUST00000115631.1 |
Diap1
|
diaphanous homolog 1 (Drosophila) |
| chr1_+_194976342 | 1.87 |
ENSMUST00000181226.1
ENSMUST00000181947.1 |
A330023F24Rik
|
RIKEN cDNA A330023F24 gene |
| chr14_+_52844465 | 1.87 |
ENSMUST00000181360.1
ENSMUST00000183652.1 |
Trav12d-1
|
T cell receptor alpha variable 12D-1 |
| chr4_-_45530330 | 1.86 |
ENSMUST00000061986.5
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr18_-_37935429 | 1.85 |
ENSMUST00000115634.1
|
Diap1
|
diaphanous homolog 1 (Drosophila) |
| chr8_+_71371283 | 1.84 |
ENSMUST00000110051.1
ENSMUST00000002469.8 ENSMUST00000110052.1 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr1_+_21218575 | 1.84 |
ENSMUST00000027065.5
ENSMUST00000027064.7 |
Tmem14a
|
transmembrane protein 14A |
| chr1_-_134234492 | 1.83 |
ENSMUST00000169927.1
|
Adora1
|
adenosine A1 receptor |
| chr2_-_73386396 | 1.83 |
ENSMUST00000112044.1
ENSMUST00000112043.1 ENSMUST00000076463.5 |
Gpr155
|
G protein-coupled receptor 155 |
| chr11_-_116138862 | 1.83 |
ENSMUST00000106439.1
|
Mrpl38
|
mitochondrial ribosomal protein L38 |
| chr7_+_44896077 | 1.80 |
ENSMUST00000071207.7
ENSMUST00000166849.1 ENSMUST00000168712.1 ENSMUST00000168389.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chr2_+_167688915 | 1.78 |
ENSMUST00000070642.3
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr4_-_89311021 | 1.77 |
ENSMUST00000097981.4
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr2_-_66256576 | 1.76 |
ENSMUST00000125446.2
ENSMUST00000102718.3 |
Ttc21b
|
tetratricopeptide repeat domain 21B |
| chr8_+_87472805 | 1.76 |
ENSMUST00000180700.2
ENSMUST00000182174.1 ENSMUST00000181159.1 |
Gm2694
|
predicted gene 2694 |
| chr10_-_78043580 | 1.75 |
ENSMUST00000145975.1
ENSMUST00000130972.1 ENSMUST00000128241.1 ENSMUST00000155021.1 ENSMUST00000140636.1 ENSMUST00000148469.1 ENSMUST00000019257.8 ENSMUST00000105395.2 ENSMUST00000156417.1 ENSMUST00000105396.2 ENSMUST00000154374.1 |
Aire
|
autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
| chr1_-_150393024 | 1.74 |
ENSMUST00000097546.2
ENSMUST00000111913.2 |
BC003331
|
cDNA sequence BC003331 |
| chr11_+_118443471 | 1.72 |
ENSMUST00000133558.1
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_+_38998302 | 1.72 |
ENSMUST00000112872.1
|
Wdr38
|
WD repeat domain 38 |
| chr18_-_3337539 | 1.71 |
ENSMUST00000142690.1
ENSMUST00000025069.4 ENSMUST00000082141.5 ENSMUST00000165086.1 ENSMUST00000149803.1 |
Crem
|
cAMP responsive element modulator |
| chr6_+_85154992 | 1.70 |
ENSMUST00000089584.5
|
Spr-ps1
|
sepiapterin reductase pseudogene 1 |
| chr11_-_53470479 | 1.70 |
ENSMUST00000057722.2
|
Gm9837
|
predicted gene 9837 |
| chr18_-_56562261 | 1.69 |
ENSMUST00000066208.6
ENSMUST00000172734.1 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr18_-_56562187 | 1.69 |
ENSMUST00000171844.2
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr5_+_42067960 | 1.68 |
ENSMUST00000087332.4
|
Gm16223
|
predicted gene 16223 |
| chr7_-_141437829 | 1.68 |
ENSMUST00000019226.7
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr1_+_58113136 | 1.67 |
ENSMUST00000040999.7
|
Aox3
|
aldehyde oxidase 3 |
| chr9_-_50528641 | 1.65 |
ENSMUST00000034570.5
|
Pts
|
6-pyruvoyl-tetrahydropterin synthase |
| chr6_-_85137743 | 1.65 |
ENSMUST00000174769.1
ENSMUST00000174286.1 ENSMUST00000045986.7 |
Spr
|
sepiapterin reductase |
| chr3_-_5576111 | 1.65 |
ENSMUST00000165309.1
ENSMUST00000164828.1 ENSMUST00000071280.5 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr8_+_87472838 | 1.65 |
ENSMUST00000180806.2
|
Gm2694
|
predicted gene 2694 |
| chr9_+_44772909 | 1.63 |
ENSMUST00000002099.3
|
Ift46
|
intraflagellar transport 46 |
| chr9_+_44772951 | 1.63 |
ENSMUST00000128150.1
|
Ift46
|
intraflagellar transport 46 |
| chr11_+_84179852 | 1.62 |
ENSMUST00000136463.2
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr14_+_52769753 | 1.60 |
ENSMUST00000178768.2
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr3_-_121171678 | 1.59 |
ENSMUST00000170781.1
ENSMUST00000039761.5 ENSMUST00000106467.1 ENSMUST00000106466.3 ENSMUST00000164925.2 |
Rwdd3
|
RWD domain containing 3 |
| chr2_-_166155624 | 1.59 |
ENSMUST00000109249.2
|
Sulf2
|
sulfatase 2 |
| chr14_-_21748610 | 1.58 |
ENSMUST00000075040.2
ENSMUST00000183943.1 |
Dusp13
|
dual specificity phosphatase 13 |
| chr7_-_141437587 | 1.58 |
ENSMUST00000172654.1
ENSMUST00000106006.1 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr13_-_41220395 | 1.58 |
ENSMUST00000021793.7
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr3_-_5576233 | 1.57 |
ENSMUST00000059021.4
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr7_-_74554726 | 1.56 |
ENSMUST00000107453.1
|
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr11_+_105975204 | 1.56 |
ENSMUST00000001964.7
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr13_-_8871751 | 1.55 |
ENSMUST00000175958.1
|
Wdr37
|
WD repeat domain 37 |
| chr18_-_56562215 | 1.55 |
ENSMUST00000170309.1
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr5_+_120649188 | 1.53 |
ENSMUST00000156722.1
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr13_+_54621801 | 1.53 |
ENSMUST00000026991.9
ENSMUST00000137413.1 ENSMUST00000135232.1 ENSMUST00000124752.1 |
Faf2
|
Fas associated factor family member 2 |
| chr9_+_110333340 | 1.53 |
ENSMUST00000098350.3
|
Scap
|
SREBF chaperone |
| chr11_-_82890541 | 1.53 |
ENSMUST00000092844.6
ENSMUST00000021033.9 ENSMUST00000018985.8 |
Rad51d
|
RAD51 homolog D |
| chr2_-_112368021 | 1.52 |
ENSMUST00000028551.3
|
Emc4
|
ER membrane protein complex subunit 4 |
| chr16_-_94313556 | 1.50 |
ENSMUST00000163193.1
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| chr6_+_108783059 | 1.49 |
ENSMUST00000032196.6
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr1_-_150392719 | 1.48 |
ENSMUST00000006167.6
ENSMUST00000094477.2 ENSMUST00000097547.3 |
BC003331
|
cDNA sequence BC003331 |
| chr14_+_52824340 | 1.48 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr15_-_76511951 | 1.47 |
ENSMUST00000023214.4
|
Dgat1
|
diacylglycerol O-acyltransferase 1 |
| chr12_+_8313425 | 1.47 |
ENSMUST00000020927.8
|
Hs1bp3
|
HCLS1 binding protein 3 |
| chr19_-_10880370 | 1.46 |
ENSMUST00000133303.1
|
Tmem109
|
transmembrane protein 109 |
| chr14_+_52753367 | 1.46 |
ENSMUST00000180717.1
ENSMUST00000183820.1 |
Trav6d-4
|
T cell receptor alpha variable 6D-4 |
| chr15_-_99087817 | 1.46 |
ENSMUST00000064462.3
|
C1ql4
|
complement component 1, q subcomponent-like 4 |
| chr7_-_4524229 | 1.45 |
ENSMUST00000154913.1
|
Tnni3
|
troponin I, cardiac 3 |
| chr11_+_50131342 | 1.44 |
ENSMUST00000093138.6
ENSMUST00000101270.4 |
Tbc1d9b
|
TBC1 domain family, member 9B |
| chr4_-_44072712 | 1.44 |
ENSMUST00000102936.2
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr11_+_104231573 | 1.44 |
ENSMUST00000132977.1
ENSMUST00000132245.1 ENSMUST00000100347.4 |
Mapt
|
microtubule-associated protein tau |
| chr14_+_53443243 | 1.43 |
ENSMUST00000177622.2
|
Trav7-3
|
T cell receptor alpha variable 7-3 |
| chr7_+_107209439 | 1.42 |
ENSMUST00000098135.1
|
Rbmxl2
|
RNA binding motif protein, X-linked-like 2 |
| chr8_-_110168204 | 1.42 |
ENSMUST00000003754.6
|
Calb2
|
calbindin 2 |
| chr14_+_53461099 | 1.41 |
ENSMUST00000181728.1
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr13_-_41220162 | 1.41 |
ENSMUST00000117096.1
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr8_-_87472576 | 1.40 |
ENSMUST00000034076.8
|
Cbln1
|
cerebellin 1 precursor protein |
| chr11_+_78178105 | 1.40 |
ENSMUST00000147819.1
|
Tlcd1
|
TLC domain containing 1 |
| chr9_-_57645561 | 1.36 |
ENSMUST00000034863.6
|
Csk
|
c-src tyrosine kinase |
| chr8_-_109962127 | 1.35 |
ENSMUST00000001722.7
ENSMUST00000051430.6 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr3_-_89160155 | 1.35 |
ENSMUST00000029686.3
|
Hcn3
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
| chr17_+_28232723 | 1.35 |
ENSMUST00000002320.8
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr14_-_55758458 | 1.33 |
ENSMUST00000001497.7
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr18_-_38211957 | 1.31 |
ENSMUST00000159405.1
ENSMUST00000160721.1 |
Pcdh1
|
protocadherin 1 |
| chr17_-_26090251 | 1.30 |
ENSMUST00000040907.6
|
Decr2
|
2-4-dienoyl-Coenzyme A reductase 2, peroxisomal |
| chr14_+_62555737 | 1.29 |
ENSMUST00000039064.7
|
Fam124a
|
family with sequence similarity 124, member A |
| chr14_+_53676141 | 1.29 |
ENSMUST00000103662.4
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr11_+_104231465 | 1.28 |
ENSMUST00000145227.1
|
Mapt
|
microtubule-associated protein tau |
| chr1_-_55363462 | 1.28 |
ENSMUST00000159398.1
|
Boll
|
bol, boule-like (Drosophila) |
| chr7_+_141455198 | 1.28 |
ENSMUST00000164016.1
ENSMUST00000064151.6 ENSMUST00000169665.1 |
Pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr18_+_74442500 | 1.28 |
ENSMUST00000074157.6
|
Myo5b
|
myosin VB |
| chr19_+_44989073 | 1.27 |
ENSMUST00000026225.8
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr11_-_76763550 | 1.25 |
ENSMUST00000010536.8
|
Gosr1
|
golgi SNAP receptor complex member 1 |
| chr7_-_45136391 | 1.25 |
ENSMUST00000146760.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr9_+_107888129 | 1.24 |
ENSMUST00000035202.2
|
Mon1a
|
MON1 homolog A (yeast) |
| chr13_+_117602439 | 1.24 |
ENSMUST00000006991.7
|
Hcn1
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 1 |
| chr5_+_140331860 | 1.22 |
ENSMUST00000071881.3
ENSMUST00000050205.5 ENSMUST00000110827.1 |
Nudt1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr14_+_52792208 | 1.20 |
ENSMUST00000178426.2
|
Trav9d-1
|
T cell receptor alpha variable 9D-1 |
| chr5_-_66618636 | 1.20 |
ENSMUST00000162382.1
ENSMUST00000160870.1 ENSMUST00000087256.5 ENSMUST00000160103.1 ENSMUST00000162349.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rad21_Smc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 4.8 | 19.2 | GO:0061625 | fructose catabolic process(GO:0006001) response to sucrose(GO:0009744) response to disaccharide(GO:0034285) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 1.4 | 4.2 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.4 | 8.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.3 | 3.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 1.2 | 4.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.2 | 4.7 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 1.1 | 3.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 1.0 | 2.9 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) thymidine metabolic process(GO:0046104) |
| 1.0 | 15.6 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.9 | 8.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.9 | 2.7 | GO:0046968 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) peptide antigen transport(GO:0046968) |
| 0.8 | 2.5 | GO:1904020 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.8 | 2.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.7 | 2.7 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.6 | 5.8 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.6 | 2.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.6 | 1.8 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.6 | 1.7 | GO:0002465 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) regulation of thymocyte migration(GO:2000410) |
| 0.5 | 1.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.5 | 7.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.5 | 1.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.5 | 1.5 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.5 | 1.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.5 | 1.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.5 | 3.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 6.8 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.4 | 2.5 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 1.3 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.4 | 1.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.4 | 1.2 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.4 | 2.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.4 | 1.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.4 | 2.3 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.4 | 1.9 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 1.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.4 | 3.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.4 | 0.7 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.4 | 2.2 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.4 | 1.4 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 | 5.4 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.3 | 1.7 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.3 | 3.6 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.3 | 3.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 2.7 | GO:0000050 | urea cycle(GO:0000050) |
| 0.3 | 1.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 1.5 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 | 4.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.3 | 1.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 0.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 1.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.3 | 1.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 1.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.2 | 3.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.7 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.2 | 1.5 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.2 | 4.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 3.7 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 1.1 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.2 | 1.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 | 1.8 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 4.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.2 | 0.9 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 1.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 1.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.9 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 2.5 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 10.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.8 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 0.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 1.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 1.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 4.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 4.5 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 11.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.2 | 2.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 1.0 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.2 | 0.6 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.2 | 0.6 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 1.4 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.8 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.1 | 2.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.4 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.6 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.3 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 2.9 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.4 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.7 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 0.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 1.8 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.5 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.2 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 11.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.7 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.1 | 0.6 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 3.8 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 1.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.8 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.6 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.1 | 3.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 4.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.8 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 2.6 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.3 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 2.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.9 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 3.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.1 | 1.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.5 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.5 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.7 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 1.9 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.3 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 8.4 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.4 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 1.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 2.0 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.1 | 1.6 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 2.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.9 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.8 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.6 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.5 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.2 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 6.0 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 2.2 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.1 | 0.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.7 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.6 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.9 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 4.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 1.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 6.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 2.7 | GO:0019217 | regulation of fatty acid metabolic process(GO:0019217) |
| 0.0 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0003352 | regulation of cilium movement(GO:0003352) protein polyglutamylation(GO:0018095) |
| 0.0 | 5.9 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.0 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.7 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 1.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.2 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.0 | 0.9 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 4.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 2.5 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 9.0 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 1.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 1.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.9 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.7 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 1.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 4.4 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 2.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:1903214 | regulation of protein targeting to mitochondrion(GO:1903214) |
| 0.0 | 0.6 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 1.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.9 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 1.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.6 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.1 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.5 | 4.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 1.4 | 7.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.8 | 2.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.6 | 5.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.5 | 3.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.5 | 2.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 3.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 1.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.4 | 2.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.3 | 1.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 2.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 1.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 1.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 1.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 2.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 3.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 2.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 1.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 5.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.5 | GO:0060473 | cortical granule(GO:0060473) |
| 0.2 | 3.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 0.5 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 4.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.7 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.1 | 2.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:1990130 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 4.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 5.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 5.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.9 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 9.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 3.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 7.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 4.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 5.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 17.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 1.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 6.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 9.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.2 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 5.5 | 16.5 | GO:0046911 | metal chelating activity(GO:0046911) |
| 3.1 | 15.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 3.0 | 8.9 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 1.7 | 8.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.5 | 4.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 1.2 | 4.9 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 1.1 | 3.4 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 1.1 | 6.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.0 | 5.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 1.0 | 3.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.0 | 4.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.8 | 5.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.7 | 2.9 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.7 | 2.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.7 | 3.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.6 | 2.5 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.6 | 7.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.6 | 8.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.6 | 2.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.6 | 1.7 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.5 | 2.7 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.5 | 2.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 5.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.5 | 1.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.5 | 2.5 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.5 | 1.5 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.5 | 1.4 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.4 | 1.3 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.4 | 1.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 2.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 0.8 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.4 | 2.9 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.4 | 3.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.4 | 1.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 3.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 1.6 | GO:0070573 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.4 | 2.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 1.7 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.3 | 2.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 1.5 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.3 | 1.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 0.8 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.3 | 2.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 3.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 3.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 5.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 2.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 1.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 2.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 2.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 4.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 1.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 2.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 11.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 2.7 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.2 | 0.5 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.2 | 4.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 0.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 2.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 1.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 0.7 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 1.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 19.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 3.0 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 0.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 3.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 3.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 2.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0032405 | exodeoxyribonuclease III activity(GO:0008853) MutLalpha complex binding(GO:0032405) |
| 0.0 | 1.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 3.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 1.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.7 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 7.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 3.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 2.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 13.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 3.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 9.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 3.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 5.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.7 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 6.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 6.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 6.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 4.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 18.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.6 | 8.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 8.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 5.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 7.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 7.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 2.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.2 | 5.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 3.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 6.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 5.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 15.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 11.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |