Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Relb
Z-value: 0.32
Transcription factors associated with Relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Relb
|
ENSMUSG00000002983.10 | avian reticuloendotheliosis viral (v-rel) oncogene related B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Relb | mm10_v2_chr7_-_19629355_19629451 | 0.11 | 5.3e-01 | Click! |
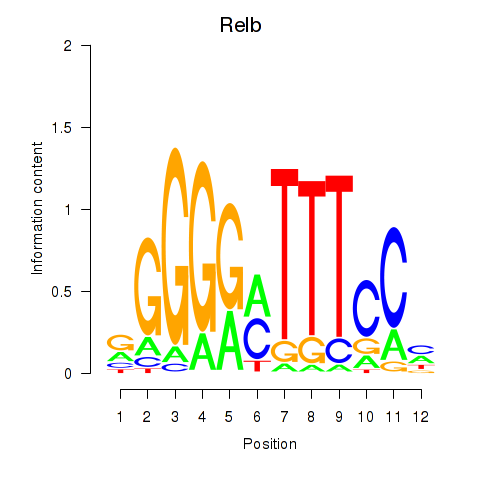
Activity profile of Relb motif
Sorted Z-values of Relb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_40089688 | 2.16 |
ENSMUST00000068094.6
ENSMUST00000080171.2 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr17_+_37193889 | 1.74 |
ENSMUST00000038844.6
|
Ubd
|
ubiquitin D |
| chr5_+_90891234 | 1.64 |
ENSMUST00000031327.8
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr1_-_171196229 | 0.97 |
ENSMUST00000111332.1
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr7_-_99626936 | 0.94 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chrX_+_36112110 | 0.72 |
ENSMUST00000033418.7
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr6_-_124741374 | 0.65 |
ENSMUST00000004389.5
|
Grcc10
|
gene rich cluster, C10 gene |
| chr15_-_31367527 | 0.64 |
ENSMUST00000076942.4
ENSMUST00000123325.1 ENSMUST00000110410.2 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr9_-_97018823 | 0.64 |
ENSMUST00000055433.4
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr2_-_5714490 | 0.63 |
ENSMUST00000044009.7
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr12_-_54862783 | 0.51 |
ENSMUST00000078124.7
|
Cfl2
|
cofilin 2, muscle |
| chr14_+_55560480 | 0.51 |
ENSMUST00000121622.1
ENSMUST00000143431.1 ENSMUST00000150481.1 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr15_-_98677451 | 0.50 |
ENSMUST00000120997.1
ENSMUST00000109149.2 ENSMUST00000003451.4 |
Rnd1
|
Rho family GTPase 1 |
| chrX_+_157698910 | 0.49 |
ENSMUST00000136141.1
|
Smpx
|
small muscle protein, X-linked |
| chr4_+_43631935 | 0.47 |
ENSMUST00000030191.8
|
Npr2
|
natriuretic peptide receptor 2 |
| chr16_-_30267524 | 0.46 |
ENSMUST00000064856.7
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chrX_+_141475385 | 0.46 |
ENSMUST00000112931.1
ENSMUST00000112930.1 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr4_-_117133953 | 0.41 |
ENSMUST00000076859.5
|
Plk3
|
polo-like kinase 3 |
| chr1_+_152399824 | 0.40 |
ENSMUST00000044311.8
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr15_-_89170688 | 0.39 |
ENSMUST00000060808.9
|
Plxnb2
|
plexin B2 |
| chr2_-_76982455 | 0.38 |
ENSMUST00000011934.5
ENSMUST00000099981.2 ENSMUST00000099980.3 ENSMUST00000111882.2 ENSMUST00000140091.1 |
Ttn
|
titin |
| chr1_+_74791516 | 0.36 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr10_+_102512216 | 0.35 |
ENSMUST00000055355.4
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr5_-_33657889 | 0.34 |
ENSMUST00000019439.7
|
Tmem129
|
transmembrane protein 129 |
| chr1_+_193301953 | 0.32 |
ENSMUST00000016315.9
|
Lamb3
|
laminin, beta 3 |
| chr1_-_135258449 | 0.32 |
ENSMUST00000003135.7
|
Elf3
|
E74-like factor 3 |
| chr10_-_77902467 | 0.32 |
ENSMUST00000057608.4
|
Lrrc3
|
leucine rich repeat containing 3 |
| chr12_-_55492587 | 0.30 |
ENSMUST00000021413.7
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr7_+_28766747 | 0.29 |
ENSMUST00000170068.1
ENSMUST00000072965.4 |
Sirt2
|
sirtuin 2 |
| chr6_+_122513583 | 0.29 |
ENSMUST00000032210.7
ENSMUST00000148517.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr11_+_90030295 | 0.27 |
ENSMUST00000092788.3
|
Tmem100
|
transmembrane protein 100 |
| chr17_+_35424842 | 0.26 |
ENSMUST00000174699.1
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr15_-_33405976 | 0.26 |
ENSMUST00000079057.6
|
1700084J12Rik
|
RIKEN cDNA 1700084J12 gene |
| chr2_+_10153563 | 0.26 |
ENSMUST00000026886.7
|
Itih5
|
inter-alpha (globulin) inhibitor H5 |
| chrX_+_157699113 | 0.26 |
ENSMUST00000112521.1
|
Smpx
|
small muscle protein, X-linked |
| chr17_+_35424870 | 0.25 |
ENSMUST00000113879.3
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr2_+_59612034 | 0.25 |
ENSMUST00000112568.1
ENSMUST00000037526.4 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_-_5063932 | 0.24 |
ENSMUST00000027986.4
|
Optn
|
optineurin |
| chr2_+_18672384 | 0.24 |
ENSMUST00000171845.1
ENSMUST00000061158.4 |
Commd3
|
COMM domain containing 3 |
| chr10_+_96616998 | 0.24 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr2_-_34958129 | 0.24 |
ENSMUST00000172159.1
|
Traf1
|
TNF receptor-associated factor 1 |
| chr7_-_19629355 | 0.24 |
ENSMUST00000049912.8
ENSMUST00000094762.3 ENSMUST00000098754.4 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chrX_+_103321398 | 0.23 |
ENSMUST00000033689.2
|
Cdx4
|
caudal type homeobox 4 |
| chr8_+_72319033 | 0.23 |
ENSMUST00000067912.7
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chr16_-_55838827 | 0.22 |
ENSMUST00000096026.2
ENSMUST00000036273.6 ENSMUST00000114457.1 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr4_-_148500449 | 0.22 |
ENSMUST00000030840.3
|
Angptl7
|
angiopoietin-like 7 |
| chr12_+_111166413 | 0.20 |
ENSMUST00000021706.4
|
Traf3
|
TNF receptor-associated factor 3 |
| chr2_-_5063996 | 0.20 |
ENSMUST00000114996.1
|
Optn
|
optineurin |
| chr18_+_37484955 | 0.20 |
ENSMUST00000053856.4
|
Pcdhb17
|
protocadherin beta 17 |
| chr7_-_70360593 | 0.19 |
ENSMUST00000032768.7
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_57149633 | 0.18 |
ENSMUST00000019633.7
|
Cd70
|
CD70 antigen |
| chr1_-_130887408 | 0.17 |
ENSMUST00000121040.1
|
Il24
|
interleukin 24 |
| chr7_-_28766469 | 0.17 |
ENSMUST00000085851.5
ENSMUST00000032815.4 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr14_+_120478443 | 0.15 |
ENSMUST00000062117.6
|
Rap2a
|
RAS related protein 2a |
| chr8_+_46739745 | 0.15 |
ENSMUST00000034041.7
|
Irf2
|
interferon regulatory factor 2 |
| chr10_-_93310963 | 0.15 |
ENSMUST00000151153.1
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr14_+_54932695 | 0.15 |
ENSMUST00000037863.5
|
Il25
|
interleukin 25 |
| chr17_+_42315947 | 0.14 |
ENSMUST00000048691.4
|
Ptchd4
|
patched domain containing 4 |
| chr11_-_23770953 | 0.13 |
ENSMUST00000102864.3
|
Rel
|
reticuloendotheliosis oncogene |
| chr14_+_62760496 | 0.13 |
ENSMUST00000181344.1
|
4931440J10Rik
|
RIKEN cDNA 4931440J10 gene |
| chr12_+_111166349 | 0.12 |
ENSMUST00000117269.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chr1_+_15287259 | 0.12 |
ENSMUST00000175681.1
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr4_-_91399984 | 0.11 |
ENSMUST00000102799.3
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr4_+_126262325 | 0.11 |
ENSMUST00000030660.8
|
Trappc3
|
trafficking protein particle complex 3 |
| chr12_+_102948843 | 0.10 |
ENSMUST00000101099.5
|
Unc79
|
unc-79 homolog (C. elegans) |
| chr2_+_119972699 | 0.10 |
ENSMUST00000066058.7
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr2_-_160313616 | 0.10 |
ENSMUST00000109475.2
|
Gm826
|
predicted gene 826 |
| chr10_-_93311073 | 0.10 |
ENSMUST00000008542.5
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr8_-_46739453 | 0.10 |
ENSMUST00000181167.1
|
Gm16675
|
predicted gene, 16675 |
| chr2_+_143546144 | 0.09 |
ENSMUST00000028905.9
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr15_-_89429093 | 0.09 |
ENSMUST00000170460.1
|
Chkb
|
choline kinase beta |
| chr17_+_35649708 | 0.08 |
ENSMUST00000171166.2
|
Sfta2
|
surfactant associated 2 |
| chr4_-_40239779 | 0.07 |
ENSMUST00000037907.6
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr2_-_27027909 | 0.07 |
ENSMUST00000102890.4
ENSMUST00000153388.1 ENSMUST00000045702.5 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr16_+_88728828 | 0.04 |
ENSMUST00000060494.6
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr4_+_99656299 | 0.04 |
ENSMUST00000087285.3
|
Foxd3
|
forkhead box D3 |
| chrX_+_163908982 | 0.04 |
ENSMUST00000069041.8
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr18_+_36559972 | 0.03 |
ENSMUST00000134146.1
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr1_-_45890078 | 0.02 |
ENSMUST00000183590.1
|
Gm5269
|
predicted gene 5269 |
| chr2_-_125506385 | 0.00 |
ENSMUST00000028633.6
|
Fbn1
|
fibrillin 1 |
| chr5_-_5663263 | 0.00 |
ENSMUST00000148193.1
|
A330021E22Rik
|
RIKEN cDNA A330021E22 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Relb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.2 | 2.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 0.7 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.2 | 0.6 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.4 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.4 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.4 | GO:0090166 | Golgi disassembly(GO:0090166) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:0097435 | fibril organization(GO:0097435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.5 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0042903 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |