Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
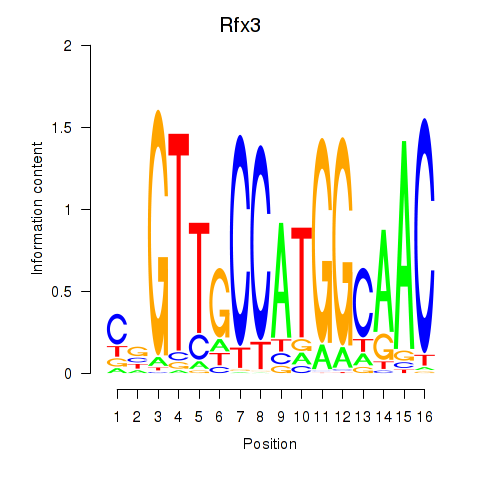
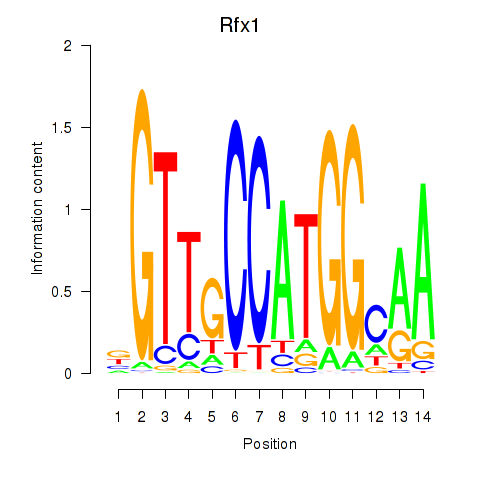
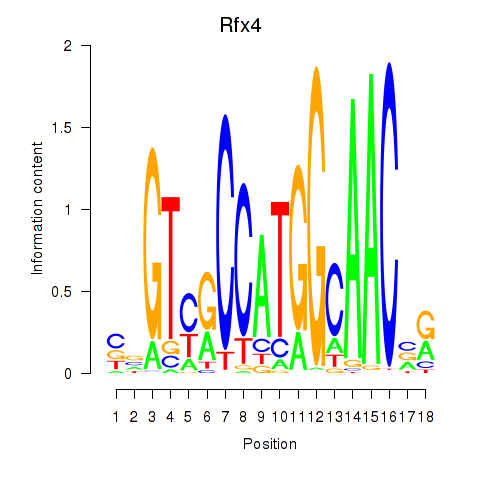
Results for Rfx3_Rfx1_Rfx4
Z-value: 1.11
Transcription factors associated with Rfx3_Rfx1_Rfx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx3
|
ENSMUSG00000040929.10 | regulatory factor X, 3 (influences HLA class II expression) |
|
Rfx1
|
ENSMUSG00000031706.6 | regulatory factor X, 1 (influences HLA class II expression) |
|
Rfx4
|
ENSMUSG00000020037.9 | regulatory factor X, 4 (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx4 | mm10_v2_chr10_+_84756055_84756084 | -0.44 | 7.1e-03 | Click! |
| Rfx1 | mm10_v2_chr8_+_84066824_84066882 | 0.37 | 2.8e-02 | Click! |
| Rfx3 | mm10_v2_chr19_-_28010995_28011054 | 0.07 | 6.7e-01 | Click! |
Activity profile of Rfx3_Rfx1_Rfx4 motif
Sorted Z-values of Rfx3_Rfx1_Rfx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_23684019 | 2.51 |
ENSMUST00000085989.5
|
Cldn9
|
claudin 9 |
| chr16_-_3908596 | 2.35 |
ENSMUST00000123235.2
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr16_+_17646564 | 1.74 |
ENSMUST00000182117.1
ENSMUST00000182671.1 ENSMUST00000182344.1 |
Ccdc74a
|
coiled-coil domain containing 74A |
| chr2_-_164745916 | 1.74 |
ENSMUST00000109328.1
ENSMUST00000043448.1 |
Wfdc3
Wfdc3
|
WAP four-disulfide core domain 3 WAP four-disulfide core domain 3 |
| chr12_-_84148449 | 1.69 |
ENSMUST00000061425.2
|
Pnma1
|
paraneoplastic antigen MA1 |
| chr12_+_113185877 | 1.62 |
ENSMUST00000058491.6
|
Tmem121
|
transmembrane protein 121 |
| chr9_+_8544196 | 1.60 |
ENSMUST00000050433.6
|
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr16_+_17646464 | 1.59 |
ENSMUST00000056962.4
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr18_+_60925612 | 1.53 |
ENSMUST00000102888.3
ENSMUST00000025519.4 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_-_71033462 | 1.50 |
ENSMUST00000156068.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr7_+_127091426 | 1.50 |
ENSMUST00000056288.5
|
AI467606
|
expressed sequence AI467606 |
| chr12_+_80518990 | 1.48 |
ENSMUST00000021558.6
|
Galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr9_+_62858085 | 1.45 |
ENSMUST00000034777.6
ENSMUST00000163820.1 |
Calml4
|
calmodulin-like 4 |
| chr1_-_90153396 | 1.44 |
ENSMUST00000113094.2
|
Iqca
|
IQ motif containing with AAA domain |
| chr17_+_56304313 | 1.40 |
ENSMUST00000113035.1
ENSMUST00000113039.2 ENSMUST00000142387.1 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr2_+_28700155 | 1.40 |
ENSMUST00000074156.6
|
Ak8
|
adenylate kinase 8 |
| chr7_-_6730412 | 1.39 |
ENSMUST00000051209.4
|
Peg3
|
paternally expressed 3 |
| chr5_+_33983534 | 1.38 |
ENSMUST00000114382.1
|
Gm1673
|
predicted gene 1673 |
| chr6_+_29985317 | 1.35 |
ENSMUST00000124522.1
ENSMUST00000141679.1 |
1700023L04Rik
|
RIKEN cDNA 1700023L04 gene |
| chr11_-_6606053 | 1.33 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
| chr19_+_47178820 | 1.32 |
ENSMUST00000111808.3
|
Neurl1a
|
neuralized homolog 1A (Drosophila) |
| chr5_+_33983437 | 1.27 |
ENSMUST00000114384.1
ENSMUST00000094869.5 ENSMUST00000114383.1 |
Gm1673
|
predicted gene 1673 |
| chr10_-_79874233 | 1.26 |
ENSMUST00000166023.1
ENSMUST00000167707.1 ENSMUST00000165601.1 |
BC005764
|
cDNA sequence BC005764 |
| chr15_-_84557776 | 1.26 |
ENSMUST00000069476.4
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like |
| chr8_+_94984399 | 1.23 |
ENSMUST00000093271.6
|
Gpr56
|
G protein-coupled receptor 56 |
| chr19_+_10001669 | 1.23 |
ENSMUST00000121418.1
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr2_-_131160006 | 1.23 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr8_+_95055094 | 1.22 |
ENSMUST00000058479.6
|
Ccdc135
|
coiled-coil domain containing 135 |
| chr9_-_61946768 | 1.20 |
ENSMUST00000034815.7
|
Kif23
|
kinesin family member 23 |
| chr2_+_166792525 | 1.20 |
ENSMUST00000065753.1
|
Trp53rk
|
transformation related protein 53 regulating kinase |
| chr5_-_114823460 | 1.13 |
ENSMUST00000140374.1
ENSMUST00000100850.4 |
Gm20499
2610524H06Rik
|
predicted gene 20499 RIKEN cDNA 2610524H06 gene |
| chr2_+_121289589 | 1.13 |
ENSMUST00000094639.3
|
Map1a
|
microtubule-associated protein 1 A |
| chr19_-_46327121 | 1.12 |
ENSMUST00000041391.4
ENSMUST00000096029.5 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr17_+_24696234 | 1.12 |
ENSMUST00000019464.7
|
Noxo1
|
NADPH oxidase organizer 1 |
| chr17_+_46254017 | 1.09 |
ENSMUST00000095262.4
|
Lrrc73
|
leucine rich repeat containing 73 |
| chr3_-_108536466 | 1.08 |
ENSMUST00000048012.6
ENSMUST00000106626.2 ENSMUST00000106625.3 |
5330417C22Rik
|
RIKEN cDNA 5330417C22 gene |
| chr13_+_91461050 | 1.07 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr7_+_99268338 | 1.07 |
ENSMUST00000107100.2
|
Map6
|
microtubule-associated protein 6 |
| chr12_+_85746539 | 1.05 |
ENSMUST00000040461.3
|
Mfsd7c
|
major facilitator superfamily domain containing 7C |
| chr5_+_108369858 | 1.05 |
ENSMUST00000100944.2
|
Gm10419
|
predicted gene 10419 |
| chr17_-_8101228 | 1.05 |
ENSMUST00000097422.4
|
Gm1604A
|
predicted gene 1604A |
| chr10_+_80295930 | 1.04 |
ENSMUST00000105359.1
|
Apc2
|
adenomatosis polyposis coli 2 |
| chr17_-_31277327 | 1.03 |
ENSMUST00000024832.7
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr10_+_42860348 | 1.03 |
ENSMUST00000063063.7
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr13_+_55464237 | 1.03 |
ENSMUST00000046533.7
|
Prr7
|
proline rich 7 (synaptic) |
| chr4_-_133967296 | 1.03 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr14_-_56102458 | 0.99 |
ENSMUST00000015583.1
|
Ctsg
|
cathepsin G |
| chr16_+_44394771 | 0.98 |
ENSMUST00000099742.2
|
Wdr52
|
WD repeat domain 52 |
| chr7_-_13054665 | 0.97 |
ENSMUST00000182515.1
ENSMUST00000069289.8 |
Mzf1
|
myeloid zinc finger 1 |
| chrX_+_7919816 | 0.96 |
ENSMUST00000041096.3
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr4_-_133967235 | 0.95 |
ENSMUST00000123234.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr7_+_80186835 | 0.95 |
ENSMUST00000107383.1
ENSMUST00000032754.7 |
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr4_-_118620763 | 0.94 |
ENSMUST00000071972.4
|
Wdr65
|
WD repeat domain 65 |
| chr8_-_4259257 | 0.93 |
ENSMUST00000053252.7
|
Ctxn1
|
cortexin 1 |
| chr14_+_60378242 | 0.92 |
ENSMUST00000022561.6
|
Amer2
|
APC membrane recruitment 2 |
| chr4_+_137681663 | 0.92 |
ENSMUST00000047243.5
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr11_+_102248842 | 0.91 |
ENSMUST00000100392.4
|
BC030867
|
cDNA sequence BC030867 |
| chr8_+_119575235 | 0.90 |
ENSMUST00000093100.2
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr2_-_73775341 | 0.90 |
ENSMUST00000112024.3
ENSMUST00000166199.1 ENSMUST00000180045.1 |
Chn1
|
chimerin (chimaerin) 1 |
| chr15_-_79062866 | 0.89 |
ENSMUST00000151889.1
ENSMUST00000040676.4 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr2_+_110017806 | 0.87 |
ENSMUST00000028580.5
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr9_+_20652107 | 0.86 |
ENSMUST00000034689.6
|
Pin1
|
protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 |
| chr5_-_114813943 | 0.86 |
ENSMUST00000061251.5
ENSMUST00000112160.3 |
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chr11_-_12026732 | 0.86 |
ENSMUST00000143915.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr5_+_30711564 | 0.85 |
ENSMUST00000114729.1
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr11_+_61505138 | 0.83 |
ENSMUST00000102657.3
|
B9d1
|
B9 protein domain 1 |
| chr2_-_30048827 | 0.82 |
ENSMUST00000113711.2
|
Wdr34
|
WD repeat domain 34 |
| chr7_-_4546567 | 0.82 |
ENSMUST00000065957.5
|
Syt5
|
synaptotagmin V |
| chr19_-_4283033 | 0.81 |
ENSMUST00000167215.1
ENSMUST00000056888.6 |
Ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr9_-_121277160 | 0.80 |
ENSMUST00000051479.6
ENSMUST00000171923.1 |
Ulk4
|
unc-51-like kinase 4 |
| chr11_-_116077927 | 0.80 |
ENSMUST00000156545.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr1_+_20951666 | 0.80 |
ENSMUST00000038447.4
|
Efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr13_-_100775844 | 0.79 |
ENSMUST00000075550.3
|
Cenph
|
centromere protein H |
| chr17_+_34604262 | 0.79 |
ENSMUST00000174041.1
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr5_+_137758133 | 0.79 |
ENSMUST00000141733.1
ENSMUST00000110985.1 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr2_+_181219998 | 0.78 |
ENSMUST00000050026.6
ENSMUST00000108835.1 |
BC051628
|
cDNA sequence BC051628 |
| chr9_-_60687459 | 0.77 |
ENSMUST00000114032.1
ENSMUST00000166168.1 ENSMUST00000132366.1 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_-_60043087 | 0.76 |
ENSMUST00000027172.6
|
Ica1l
|
islet cell autoantigen 1-like |
| chr8_-_84197667 | 0.76 |
ENSMUST00000181282.1
|
Gm26887
|
predicted gene, 26887 |
| chr10_+_42860648 | 0.75 |
ENSMUST00000105495.1
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr12_-_110840905 | 0.75 |
ENSMUST00000177224.1
ENSMUST00000084974.4 ENSMUST00000070565.8 |
Stk30
|
serine/threonine kinase 30 |
| chr4_-_127330799 | 0.73 |
ENSMUST00000046532.3
|
Gjb3
|
gap junction protein, beta 3 |
| chr3_-_89245159 | 0.73 |
ENSMUST00000090924.6
|
Trim46
|
tripartite motif-containing 46 |
| chr17_+_35049966 | 0.73 |
ENSMUST00000007257.9
|
Clic1
|
chloride intracellular channel 1 |
| chr1_+_172521044 | 0.72 |
ENSMUST00000085894.5
ENSMUST00000161140.1 ENSMUST00000162988.1 |
Ccdc19
|
coiled-coil domain containing 19 |
| chr5_-_138172383 | 0.72 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr11_-_116077954 | 0.71 |
ENSMUST00000106451.1
ENSMUST00000075036.2 |
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr13_+_21945084 | 0.70 |
ENSMUST00000176511.1
ENSMUST00000102978.1 ENSMUST00000152258.2 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr1_+_171767123 | 0.70 |
ENSMUST00000015460.4
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr11_+_106374820 | 0.70 |
ENSMUST00000106816.1
|
2310007L24Rik
|
RIKEN cDNA 2310007L24 gene |
| chr2_-_131174653 | 0.69 |
ENSMUST00000127987.1
|
Spef1
|
sperm flagellar 1 |
| chr3_+_67374116 | 0.69 |
ENSMUST00000061322.8
|
Mlf1
|
myeloid leukemia factor 1 |
| chr6_-_102464667 | 0.68 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr9_-_27155418 | 0.68 |
ENSMUST00000167074.1
ENSMUST00000034472.8 |
Jam3
|
junction adhesion molecule 3 |
| chr3_+_124321031 | 0.68 |
ENSMUST00000058994.4
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr3_+_67374091 | 0.67 |
ENSMUST00000077916.5
|
Mlf1
|
myeloid leukemia factor 1 |
| chr5_+_124598749 | 0.67 |
ENSMUST00000130912.1
ENSMUST00000100706.3 |
Tctn2
|
tectonic family member 2 |
| chr5_-_136170634 | 0.67 |
ENSMUST00000041048.1
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr7_-_13054514 | 0.67 |
ENSMUST00000182087.1
|
Mzf1
|
myeloid zinc finger 1 |
| chr11_-_87359011 | 0.67 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr5_+_77016023 | 0.67 |
ENSMUST00000031161.4
ENSMUST00000117880.1 |
1700023E05Rik
|
RIKEN cDNA 1700023E05 gene |
| chr7_+_118855735 | 0.66 |
ENSMUST00000098087.2
ENSMUST00000106547.1 |
Iqck
|
IQ motif containing K |
| chr2_+_110017879 | 0.66 |
ENSMUST00000150183.2
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr5_-_140702241 | 0.65 |
ENSMUST00000077890.5
ENSMUST00000041783.7 ENSMUST00000142081.1 |
Iqce
|
IQ motif containing E |
| chr17_+_12119274 | 0.65 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr1_-_156674290 | 0.64 |
ENSMUST00000079625.4
|
Tor3a
|
torsin family 3, member A |
| chr14_-_49245389 | 0.64 |
ENSMUST00000130853.1
ENSMUST00000022398.7 |
1700011H14Rik
|
RIKEN cDNA 1700011H14 gene |
| chrX_+_73123068 | 0.63 |
ENSMUST00000179117.1
|
Gm14685
|
predicted gene 14685 |
| chr11_+_87760533 | 0.63 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr17_-_46247968 | 0.62 |
ENSMUST00000142706.2
ENSMUST00000173349.1 ENSMUST00000087026.6 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr11_-_116077562 | 0.61 |
ENSMUST00000174822.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr3_-_55055038 | 0.61 |
ENSMUST00000029368.2
|
Ccna1
|
cyclin A1 |
| chr18_-_77047243 | 0.61 |
ENSMUST00000137354.1
ENSMUST00000137498.1 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr6_-_122339627 | 0.61 |
ENSMUST00000161210.1
ENSMUST00000161054.1 ENSMUST00000159252.1 ENSMUST00000161739.1 ENSMUST00000161149.1 |
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr11_+_49794157 | 0.60 |
ENSMUST00000020629.4
|
Gfpt2
|
glutamine fructose-6-phosphate transaminase 2 |
| chr11_-_67965631 | 0.60 |
ENSMUST00000021287.5
ENSMUST00000126766.1 |
Wdr16
|
WD repeat domain 16 |
| chr11_+_43474276 | 0.60 |
ENSMUST00000173002.1
ENSMUST00000057679.3 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr4_+_126058557 | 0.60 |
ENSMUST00000035497.4
|
Oscp1
|
organic solute carrier partner 1 |
| chr17_-_31855782 | 0.59 |
ENSMUST00000024839.4
|
Sik1
|
salt inducible kinase 1 |
| chr9_+_107547288 | 0.59 |
ENSMUST00000010188.7
|
Zmynd10
|
zinc finger, MYND domain containing 10 |
| chr8_+_69832633 | 0.58 |
ENSMUST00000131637.2
ENSMUST00000081503.6 |
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr5_-_115300912 | 0.58 |
ENSMUST00000112090.1
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr8_-_84104773 | 0.58 |
ENSMUST00000041367.7
|
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr7_+_126950837 | 0.58 |
ENSMUST00000106332.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr8_-_22125030 | 0.58 |
ENSMUST00000169834.1
|
Nek5
|
NIMA (never in mitosis gene a)-related expressed kinase 5 |
| chr7_-_127588595 | 0.57 |
ENSMUST00000072155.3
|
Gm166
|
predicted gene 166 |
| chr11_+_69070790 | 0.57 |
ENSMUST00000075980.5
ENSMUST00000094081.4 |
Tmem107
|
transmembrane protein 107 |
| chr7_-_140082246 | 0.57 |
ENSMUST00000166758.2
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr17_+_84626458 | 0.57 |
ENSMUST00000025101.8
|
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr6_-_146954399 | 0.57 |
ENSMUST00000111622.1
ENSMUST00000036592.8 |
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene |
| chr5_-_115300957 | 0.56 |
ENSMUST00000009157.3
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr15_+_54745702 | 0.56 |
ENSMUST00000050027.8
|
Nov
|
nephroblastoma overexpressed gene |
| chr7_+_4740111 | 0.56 |
ENSMUST00000098853.2
|
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr7_+_4740178 | 0.56 |
ENSMUST00000108583.2
|
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr11_+_75999912 | 0.56 |
ENSMUST00000066408.5
|
1700016K19Rik
|
RIKEN cDNA 1700016K19 gene |
| chr9_-_98563580 | 0.56 |
ENSMUST00000058992.2
|
4930579K19Rik
|
RIKEN cDNA 4930579K19 gene |
| chrX_+_136993147 | 0.56 |
ENSMUST00000113067.1
ENSMUST00000101227.2 |
Zcchc18
|
zinc finger, CCHC domain containing 18 |
| chr7_+_16816299 | 0.55 |
ENSMUST00000108495.2
|
Strn4
|
striatin, calmodulin binding protein 4 |
| chr11_-_3895085 | 0.55 |
ENSMUST00000020712.4
|
4921536K21Rik
|
RIKEN cDNA 4921536K21 gene |
| chr9_-_8042785 | 0.55 |
ENSMUST00000065291.1
|
9230110C19Rik
|
RIKEN cDNA 9230110C19 gene |
| chr5_-_115652974 | 0.55 |
ENSMUST00000121746.1
ENSMUST00000118576.1 |
Ccdc64
|
coiled-coil domain containing 64 |
| chr8_-_111933761 | 0.55 |
ENSMUST00000034429.7
|
Tmem231
|
transmembrane protein 231 |
| chr7_-_31051431 | 0.54 |
ENSMUST00000073892.4
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr15_-_68258787 | 0.54 |
ENSMUST00000162173.1
ENSMUST00000160248.1 ENSMUST00000159430.1 ENSMUST00000162054.1 |
Zfat
|
zinc finger and AT hook domain containing |
| chr14_+_77156733 | 0.54 |
ENSMUST00000022589.7
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr10_+_122678764 | 0.54 |
ENSMUST00000161487.1
ENSMUST00000067918.5 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr4_+_111720187 | 0.54 |
ENSMUST00000084354.3
|
Spata6
|
spermatogenesis associated 6 |
| chr7_+_126950518 | 0.54 |
ENSMUST00000106335.1
ENSMUST00000146017.1 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr16_-_16829276 | 0.54 |
ENSMUST00000023468.5
|
Spag6
|
sperm associated antigen 6 |
| chr17_-_80207299 | 0.53 |
ENSMUST00000063417.9
|
Srsf7
|
serine/arginine-rich splicing factor 7 |
| chr8_-_92355764 | 0.53 |
ENSMUST00000180102.1
ENSMUST00000179421.1 ENSMUST00000179222.1 ENSMUST00000179029.1 |
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr3_-_89245005 | 0.53 |
ENSMUST00000107464.1
|
Trim46
|
tripartite motif-containing 46 |
| chr15_+_79141324 | 0.52 |
ENSMUST00000040077.6
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr7_-_44748306 | 0.52 |
ENSMUST00000118162.1
ENSMUST00000140599.2 ENSMUST00000120798.1 |
Zfp473
|
zinc finger protein 473 |
| chr4_+_111719975 | 0.52 |
ENSMUST00000038868.7
ENSMUST00000070513.6 ENSMUST00000153746.1 |
Spata6
|
spermatogenesis associated 6 |
| chr15_+_75704280 | 0.52 |
ENSMUST00000121137.1
ENSMUST00000023244.5 |
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr9_-_60688118 | 0.52 |
ENSMUST00000114034.2
ENSMUST00000065603.5 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_-_152766281 | 0.52 |
ENSMUST00000111859.1
ENSMUST00000148865.1 |
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr4_-_128962420 | 0.52 |
ENSMUST00000119354.1
ENSMUST00000106068.1 ENSMUST00000030581.3 |
Adc
|
arginine decarboxylase |
| chr10_-_68541842 | 0.52 |
ENSMUST00000020103.2
|
1700040L02Rik
|
RIKEN cDNA 1700040L02 gene |
| chr4_-_138725262 | 0.52 |
ENSMUST00000105811.2
|
Ubxn10
|
UBX domain protein 10 |
| chr8_-_119575143 | 0.52 |
ENSMUST00000036049.4
|
Hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr14_-_60197173 | 0.52 |
ENSMUST00000131670.1
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr3_-_101287897 | 0.51 |
ENSMUST00000029456.4
|
Cd2
|
CD2 antigen |
| chr17_-_6655939 | 0.51 |
ENSMUST00000179554.1
|
Dynlt1f
|
dynein light chain Tctex-type 1F |
| chr6_+_128375456 | 0.51 |
ENSMUST00000100926.2
|
4933413G19Rik
|
RIKEN cDNA 4933413G19 gene |
| chr18_+_31609512 | 0.51 |
ENSMUST00000164667.1
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr5_+_30711849 | 0.51 |
ENSMUST00000088081.4
ENSMUST00000101442.3 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr7_+_126950687 | 0.51 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr4_+_107830958 | 0.50 |
ENSMUST00000106731.2
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chrX_+_74837196 | 0.50 |
ENSMUST00000114116.1
|
Gm5936
|
predicted gene 5936 |
| chr12_+_52699297 | 0.50 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr11_-_101551837 | 0.50 |
ENSMUST00000017290.4
|
Brca1
|
breast cancer 1 |
| chr11_-_114795888 | 0.49 |
ENSMUST00000000206.3
|
Btbd17
|
BTB (POZ) domain containing 17 |
| chr4_-_154042660 | 0.49 |
ENSMUST00000047207.6
|
Ccdc27
|
coiled-coil domain containing 27 |
| chr6_+_119175247 | 0.49 |
ENSMUST00000112777.2
ENSMUST00000073909.5 |
Dcp1b
|
DCP1 decapping enzyme homolog B (S. cerevisiae) |
| chr17_+_6430112 | 0.49 |
ENSMUST00000179569.1
|
Dynlt1b
|
dynein light chain Tctex-type 1B |
| chr11_-_118342500 | 0.49 |
ENSMUST00000103024.3
|
BC100451
|
cDNA sequence BC100451 |
| chr10_+_20952547 | 0.49 |
ENSMUST00000105525.4
|
Ahi1
|
Abelson helper integration site 1 |
| chr2_+_153492790 | 0.49 |
ENSMUST00000109783.1
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr7_+_4740137 | 0.48 |
ENSMUST00000130215.1
ENSMUST00000108582.3 |
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr10_+_119992962 | 0.48 |
ENSMUST00000154238.1
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr10_-_39133914 | 0.48 |
ENSMUST00000135785.1
|
Fam229b
|
family with sequence similarity 229, member B |
| chr19_-_7241216 | 0.48 |
ENSMUST00000025675.9
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr16_+_20696175 | 0.48 |
ENSMUST00000128273.1
|
Fam131a
|
family with sequence similarity 131, member A |
| chr7_+_139212974 | 0.48 |
ENSMUST00000016124.8
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr2_-_21205342 | 0.48 |
ENSMUST00000027992.2
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr10_+_119992916 | 0.48 |
ENSMUST00000105261.2
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr3_-_101287879 | 0.48 |
ENSMUST00000152321.1
|
Cd2
|
CD2 antigen |
| chr11_-_119300070 | 0.48 |
ENSMUST00000026667.8
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr7_+_19149722 | 0.47 |
ENSMUST00000049294.2
|
Snrpd2
|
small nuclear ribonucleoprotein D2 |
| chr19_-_10457447 | 0.47 |
ENSMUST00000171400.2
|
Lrrc10b
|
leucine rich repeat containing 10B |
| chr15_-_79141197 | 0.47 |
ENSMUST00000169604.1
|
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr4_-_152477433 | 0.47 |
ENSMUST00000159186.1
ENSMUST00000162017.1 ENSMUST00000030768.2 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chrX_-_136993027 | 0.47 |
ENSMUST00000171738.1
ENSMUST00000056674.5 ENSMUST00000129807.1 |
Slc25a53
|
solute carrier family 25, member 53 |
| chr5_+_103425181 | 0.47 |
ENSMUST00000048957.9
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr17_+_6007580 | 0.47 |
ENSMUST00000115784.1
ENSMUST00000115785.1 |
Synj2
|
synaptojanin 2 |
| chr17_-_34836869 | 0.47 |
ENSMUST00000077477.5
|
Stk19
|
serine/threonine kinase 19 |
| chr12_-_69790660 | 0.47 |
ENSMUST00000021377.4
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr5_+_111581422 | 0.46 |
ENSMUST00000064930.3
|
C130026L21Rik
|
RIKEN cDNA C130026L21 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx3_Rfx1_Rfx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 0.7 | GO:2000349 | negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.4 | 1.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 1.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 0.8 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.2 | 5.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 2.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.2 | 0.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 0.7 | GO:0045763 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.2 | 0.6 | GO:1904057 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) negative regulation of sensory perception of pain(GO:1904057) bone regeneration(GO:1990523) |
| 0.2 | 0.6 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 0.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 0.5 | GO:0030862 | positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 | 0.2 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 1.6 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.5 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 0.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 1.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 1.0 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.4 | GO:1903898 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.6 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 1.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 1.4 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 1.9 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 0.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 1.0 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.1 | 0.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 1.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.7 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.5 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.3 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 1.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 2.0 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 0.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.2 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.2 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.1 | 0.2 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.8 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.4 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.2 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.2 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.9 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.8 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 1.2 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.1 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 1.0 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 1.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0042697 | menopause(GO:0042697) |
| 0.0 | 0.5 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.0 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 1.4 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.3 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.0 | 0.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.5 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 1.5 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.2 | 1.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 3.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 2.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 0.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.3 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 1.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.3 | GO:0016342 | catenin complex(GO:0016342) lamellipodium membrane(GO:0031258) |
| 0.1 | 1.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 1.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.3 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 4.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 4.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 0.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 1.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 1.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 2.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 0.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.8 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.5 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 1.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.8 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.2 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.5 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 1.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 1.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.3 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.2 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 1.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 2.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:1990599 | oxidized DNA binding(GO:0032356) 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.3 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |