Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
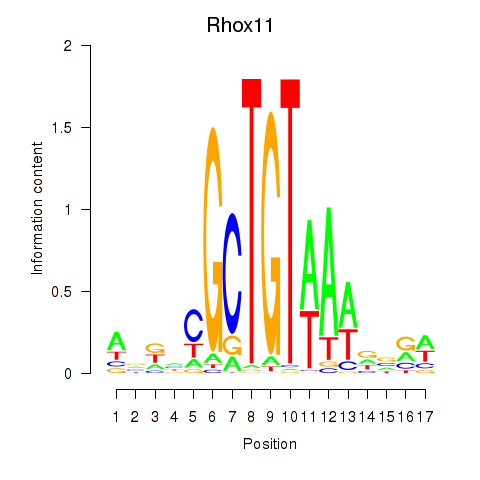
Results for Rhox11
Z-value: 0.66
Transcription factors associated with Rhox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rhox11
|
ENSMUSG00000051038.9 | reproductive homeobox 11 |
Activity profile of Rhox11 motif
Sorted Z-values of Rhox11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_122100951 | 5.04 |
ENSMUST00000014080.6
ENSMUST00000111750.1 ENSMUST00000139213.1 ENSMUST00000111751.1 ENSMUST00000155612.1 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr7_-_142699510 | 3.33 |
ENSMUST00000105934.1
|
Ins2
|
insulin II |
| chr3_-_123034943 | 2.43 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr7_-_28008416 | 1.80 |
ENSMUST00000180024.1
|
Zfp850
|
zinc finger protein 850 |
| chr1_-_175688353 | 1.75 |
ENSMUST00000104984.1
|
Chml
|
choroideremia-like |
| chr11_-_102082464 | 1.66 |
ENSMUST00000100398.4
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr17_+_40811089 | 1.65 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr11_+_70639118 | 1.56 |
ENSMUST00000055184.6
ENSMUST00000108551.2 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr1_+_40805578 | 1.54 |
ENSMUST00000114765.2
|
Tmem182
|
transmembrane protein 182 |
| chr10_+_58394361 | 1.53 |
ENSMUST00000020077.4
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr9_-_60838200 | 1.51 |
ENSMUST00000063858.7
|
Gm9869
|
predicted gene 9869 |
| chr10_+_58394381 | 1.51 |
ENSMUST00000105468.1
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_+_126556994 | 1.44 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr6_+_78405148 | 1.42 |
ENSMUST00000023906.2
|
Reg2
|
regenerating islet-derived 2 |
| chr4_+_126556935 | 1.23 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr7_-_119479249 | 1.19 |
ENSMUST00000033263.4
|
Umod
|
uromodulin |
| chr17_-_47016956 | 1.12 |
ENSMUST00000165525.1
|
Gm16494
|
predicted gene 16494 |
| chr11_-_30198232 | 1.06 |
ENSMUST00000102838.3
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr11_+_11487671 | 1.04 |
ENSMUST00000020410.4
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chr5_+_17574268 | 1.01 |
ENSMUST00000030568.7
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr14_-_20496780 | 0.94 |
ENSMUST00000022353.3
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr6_-_129660556 | 0.94 |
ENSMUST00000119533.1
ENSMUST00000145984.1 ENSMUST00000118401.1 ENSMUST00000071920.4 |
Klrc2
|
killer cell lectin-like receptor subfamily C, member 2 |
| chr17_-_37280418 | 0.94 |
ENSMUST00000077585.2
|
Olfr99
|
olfactory receptor 99 |
| chr18_-_60501983 | 0.93 |
ENSMUST00000042710.6
|
Smim3
|
small integral membrane protein 3 |
| chr8_+_105827721 | 0.93 |
ENSMUST00000034365.4
|
Tsnaxip1
|
translin-associated factor X (Tsnax) interacting protein 1 |
| chrX_-_102157065 | 0.92 |
ENSMUST00000056904.2
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr5_+_43818893 | 0.92 |
ENSMUST00000101237.4
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr5_-_139130159 | 0.90 |
ENSMUST00000129851.1
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr5_+_17574726 | 0.87 |
ENSMUST00000169603.1
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr12_+_74288735 | 0.84 |
ENSMUST00000095617.1
|
1700086L19Rik
|
RIKEN cDNA 1700086L19 gene |
| chr5_+_37735519 | 0.79 |
ENSMUST00000073554.3
|
Cytl1
|
cytokine-like 1 |
| chr1_-_153186447 | 0.73 |
ENSMUST00000027753.6
|
Lamc2
|
laminin, gamma 2 |
| chr9_+_86743616 | 0.71 |
ENSMUST00000036426.6
|
Prss35
|
protease, serine, 35 |
| chr11_+_29373618 | 0.71 |
ENSMUST00000040182.6
ENSMUST00000109477.1 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr4_+_62480025 | 0.70 |
ENSMUST00000030088.5
ENSMUST00000107449.3 |
Bspry
|
B-box and SPRY domain containing |
| chr13_-_95525239 | 0.68 |
ENSMUST00000022185.8
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr12_+_104101087 | 0.68 |
ENSMUST00000021495.3
|
Serpina5
|
serine (or cysteine) peptidase inhibitor, clade A, member 5 |
| chr7_-_46795661 | 0.66 |
ENSMUST00000123725.1
|
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr5_+_143933059 | 0.61 |
ENSMUST00000166847.1
|
Rsph10b
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr2_-_5862923 | 0.59 |
ENSMUST00000071016.2
|
Gm13199
|
predicted gene 13199 |
| chr10_-_61273242 | 0.58 |
ENSMUST00000120336.1
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr15_+_25742314 | 0.56 |
ENSMUST00000135981.1
|
Myo10
|
myosin X |
| chr9_+_86743641 | 0.56 |
ENSMUST00000179574.1
|
Prss35
|
protease, serine, 35 |
| chr12_-_99883429 | 0.54 |
ENSMUST00000046485.3
|
Efcab11
|
EF-hand calcium binding domain 11 |
| chr9_-_20644726 | 0.53 |
ENSMUST00000148631.1
ENSMUST00000131128.1 ENSMUST00000151861.1 ENSMUST00000131343.1 ENSMUST00000086458.3 |
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr13_-_3611064 | 0.51 |
ENSMUST00000096069.3
|
BC016423
|
cDNA sequence BC016423 |
| chr11_-_100355383 | 0.49 |
ENSMUST00000146878.2
|
Hap1
|
huntingtin-associated protein 1 |
| chr2_-_150362708 | 0.49 |
ENSMUST00000051153.5
|
3300002I08Rik
|
RIKEN cDNA 3300002I08 gene |
| chr9_-_8134294 | 0.48 |
ENSMUST00000037397.6
|
AK129341
|
cDNA sequence AK129341 |
| chr13_-_23562369 | 0.42 |
ENSMUST00000105107.1
|
Hist1h3e
|
histone cluster 1, H3e |
| chr6_-_129451906 | 0.40 |
ENSMUST00000037481.7
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr11_-_97700327 | 0.39 |
ENSMUST00000018681.7
|
Pcgf2
|
polycomb group ring finger 2 |
| chr5_+_121220191 | 0.39 |
ENSMUST00000119892.2
ENSMUST00000042614.6 |
Gm15800
|
predicted gene 15800 |
| chr18_+_62977854 | 0.35 |
ENSMUST00000150267.1
|
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr19_-_10974664 | 0.35 |
ENSMUST00000072748.6
|
Ms4a10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr9_-_67043709 | 0.34 |
ENSMUST00000113689.1
ENSMUST00000113684.1 |
Tpm1
|
tropomyosin 1, alpha |
| chr3_-_92485886 | 0.34 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr10_-_61273409 | 0.32 |
ENSMUST00000092486.4
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr8_-_105827190 | 0.32 |
ENSMUST00000041400.5
|
Ranbp10
|
RAN binding protein 10 |
| chr7_+_126584937 | 0.31 |
ENSMUST00000039522.6
|
Apobr
|
apolipoprotein B receptor |
| chr1_+_87853264 | 0.31 |
ENSMUST00000027517.7
|
Dgkd
|
diacylglycerol kinase, delta |
| chr3_-_121171678 | 0.30 |
ENSMUST00000170781.1
ENSMUST00000039761.5 ENSMUST00000106467.1 ENSMUST00000106466.3 ENSMUST00000164925.2 |
Rwdd3
|
RWD domain containing 3 |
| chr6_+_135065651 | 0.30 |
ENSMUST00000050104.7
|
Gprc5a
|
G protein-coupled receptor, family C, group 5, member A |
| chr15_+_36179299 | 0.29 |
ENSMUST00000047348.3
|
Spag1
|
sperm associated antigen 1 |
| chr3_+_90603767 | 0.28 |
ENSMUST00000001046.5
ENSMUST00000107330.1 |
S100a4
|
S100 calcium binding protein A4 |
| chr11_-_69895523 | 0.28 |
ENSMUST00000001631.6
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr5_-_114858682 | 0.28 |
ENSMUST00000066959.4
|
Gm9936
|
predicted gene 9936 |
| chr13_+_95325195 | 0.26 |
ENSMUST00000045909.7
|
Zbed3
|
zinc finger, BED domain containing 3 |
| chr10_+_19712587 | 0.26 |
ENSMUST00000020185.3
|
Il20ra
|
interleukin 20 receptor, alpha |
| chr9_-_107289847 | 0.26 |
ENSMUST00000035194.2
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr19_-_8818924 | 0.25 |
ENSMUST00000153281.1
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr12_+_76837408 | 0.23 |
ENSMUST00000041008.9
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr9_-_20521375 | 0.23 |
ENSMUST00000068296.7
ENSMUST00000174462.1 |
Zfp266
|
zinc finger protein 266 |
| chr6_-_34977999 | 0.19 |
ENSMUST00000044387.7
|
2010107G12Rik
|
RIKEN cDNA 2010107G12 gene |
| chr14_-_46822232 | 0.19 |
ENSMUST00000111817.1
ENSMUST00000079314.5 |
Gmfb
|
glia maturation factor, beta |
| chr13_+_49653297 | 0.17 |
ENSMUST00000021824.7
|
Nol8
|
nucleolar protein 8 |
| chr11_-_5381734 | 0.16 |
ENSMUST00000172492.1
|
Znrf3
|
zinc and ring finger 3 |
| chr7_-_14123042 | 0.15 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr7_+_5051515 | 0.15 |
ENSMUST00000069324.5
|
Zfp580
|
zinc finger protein 580 |
| chr3_-_144975011 | 0.14 |
ENSMUST00000075496.4
ENSMUST00000029923.6 |
Clca6
|
chloride channel calcium activated 6 |
| chrX_+_105079735 | 0.14 |
ENSMUST00000033577.4
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr11_-_97782377 | 0.12 |
ENSMUST00000128801.1
|
Rpl23
|
ribosomal protein L23 |
| chr8_-_81014866 | 0.12 |
ENSMUST00000042724.6
|
Usp38
|
ubiquitin specific peptidase 38 |
| chr11_-_97782409 | 0.12 |
ENSMUST00000103146.4
|
Rpl23
|
ribosomal protein L23 |
| chr1_-_44061936 | 0.10 |
ENSMUST00000168641.1
|
Gm8251
|
predicted gene 8251 |
| chr3_+_127791374 | 0.10 |
ENSMUST00000171621.1
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr3_+_68468162 | 0.08 |
ENSMUST00000182532.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr11_-_102880981 | 0.08 |
ENSMUST00000107060.1
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr11_-_102880925 | 0.08 |
ENSMUST00000021306.7
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr13_+_34924409 | 0.06 |
ENSMUST00000160279.1
|
4933417A18Rik
|
RIKEN cDNA 4933417A18 gene |
| chr2_-_131352857 | 0.06 |
ENSMUST00000059372.4
|
Rnf24
|
ring finger protein 24 |
| chr10_-_59951753 | 0.05 |
ENSMUST00000020308.3
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr9_-_110654161 | 0.05 |
ENSMUST00000133191.1
ENSMUST00000167320.1 |
Nbeal2
|
neurobeachin-like 2 |
| chr3_+_156561792 | 0.05 |
ENSMUST00000074015.4
|
Negr1
|
neuronal growth regulator 1 |
| chr9_-_78489141 | 0.05 |
ENSMUST00000154207.1
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr10_+_21295979 | 0.03 |
ENSMUST00000020153.8
ENSMUST00000092674.6 |
Hbs1l
|
Hbs1-like (S. cerevisiae) |
| chr2_-_57113053 | 0.03 |
ENSMUST00000112627.1
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr13_-_71963713 | 0.03 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr11_+_74082907 | 0.03 |
ENSMUST00000178159.1
|
Zfp616
|
zinc finger protein 616 |
| chr5_+_138187485 | 0.03 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr1_+_180942325 | 0.02 |
ENSMUST00000161847.1
|
Tmem63a
|
transmembrane protein 63a |
| chr1_+_24177610 | 0.02 |
ENSMUST00000054588.8
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr7_+_46983593 | 0.02 |
ENSMUST00000070660.4
|
Gm9999
|
predicted gene 9999 |
| chr7_-_46795881 | 0.01 |
ENSMUST00000107653.1
ENSMUST00000107654.1 ENSMUST00000014562.7 ENSMUST00000152759.1 |
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr7_-_119720742 | 0.00 |
ENSMUST00000033236.7
|
Thumpd1
|
THUMP domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.7 | 5.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.5 | 1.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 3.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 1.2 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.3 | 1.9 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 2.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 0.7 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 1.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.5 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.1 | 1.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.9 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 2.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.9 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 1.7 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 1.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 0.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 1.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.7 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 3.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 0.7 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 1.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0042627 | low-density lipoprotein particle(GO:0034362) chylomicron(GO:0042627) |
| 0.0 | 2.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 2.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 0.9 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.2 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 5.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 3.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.6 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 3.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 5.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |