Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
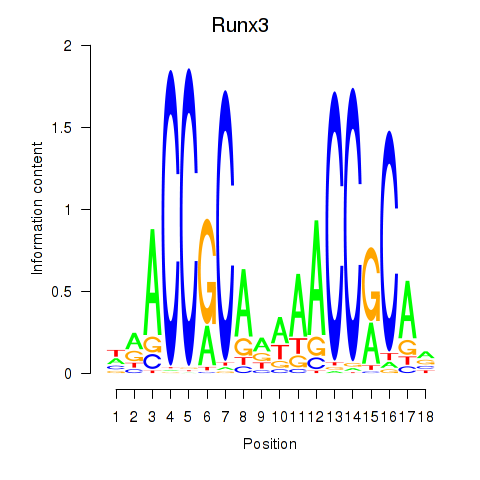
Results for Runx3
Z-value: 0.70
Transcription factors associated with Runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx3
|
ENSMUSG00000070691.4 | runt related transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx3 | mm10_v2_chr4_+_135120640_135120660 | 0.52 | 1.1e-03 | Click! |
Activity profile of Runx3 motif
Sorted Z-values of Runx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23551648 | 4.23 |
ENSMUST00000102971.1
|
Hist1h4f
|
histone cluster 1, H4f |
| chr11_-_87875524 | 3.07 |
ENSMUST00000049768.3
|
Epx
|
eosinophil peroxidase |
| chr15_-_78855517 | 2.98 |
ENSMUST00000044584.4
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr7_-_142095266 | 2.95 |
ENSMUST00000039926.3
|
Dusp8
|
dual specificity phosphatase 8 |
| chr13_-_22041352 | 2.88 |
ENSMUST00000102977.2
|
Hist1h4i
|
histone cluster 1, H4i |
| chr11_-_100207507 | 2.75 |
ENSMUST00000007272.7
|
Krt14
|
keratin 14 |
| chr9_-_32928928 | 2.74 |
ENSMUST00000185169.1
|
RP24-308I2.1
|
RP24-308I2.1 |
| chr7_+_127091426 | 2.71 |
ENSMUST00000056288.5
|
AI467606
|
expressed sequence AI467606 |
| chr13_-_23698454 | 2.18 |
ENSMUST00000102967.1
|
Hist1h4c
|
histone cluster 1, H4c |
| chr2_+_4017727 | 2.11 |
ENSMUST00000177457.1
|
Frmd4a
|
FERM domain containing 4A |
| chr13_-_51734695 | 1.87 |
ENSMUST00000110039.1
|
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr17_+_27556641 | 1.71 |
ENSMUST00000119486.1
ENSMUST00000118599.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr7_-_67803489 | 1.70 |
ENSMUST00000181235.1
|
4833412C05Rik
|
RIKEN cDNA 4833412C05 gene |
| chr10_+_76531593 | 1.65 |
ENSMUST00000048678.6
|
Lss
|
lanosterol synthase |
| chr13_-_21832194 | 1.65 |
ENSMUST00000102979.1
|
Hist1h4n
|
histone cluster 1, H4n |
| chr17_+_27556668 | 1.61 |
ENSMUST00000117254.1
ENSMUST00000118570.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr7_+_35802593 | 1.58 |
ENSMUST00000052454.2
|
E130304I02Rik
|
RIKEN cDNA E130304I02 gene |
| chr15_-_101850778 | 1.53 |
ENSMUST00000023790.3
|
Krt1
|
keratin 1 |
| chr17_+_27556613 | 1.47 |
ENSMUST00000117600.1
ENSMUST00000114888.3 |
Hmga1
|
high mobility group AT-hook 1 |
| chr4_-_120570252 | 1.45 |
ENSMUST00000030381.7
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chr13_-_59769751 | 1.42 |
ENSMUST00000057115.6
|
Isca1
|
iron-sulfur cluster assembly 1 homolog (S. cerevisiae) |
| chr13_-_76018524 | 1.32 |
ENSMUST00000050997.1
ENSMUST00000179078.1 ENSMUST00000167271.1 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr11_-_86357570 | 1.30 |
ENSMUST00000043624.8
|
Med13
|
mediator complex subunit 13 |
| chr12_+_26469204 | 1.26 |
ENSMUST00000020969.3
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr1_+_74278550 | 1.22 |
ENSMUST00000077985.3
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr11_-_70220794 | 1.21 |
ENSMUST00000159867.1
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr11_-_70220776 | 1.20 |
ENSMUST00000141290.1
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr9_-_108094459 | 1.14 |
ENSMUST00000081309.7
|
Apeh
|
acylpeptide hydrolase |
| chr11_-_5878207 | 1.12 |
ENSMUST00000102922.3
|
Pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr6_-_136804414 | 1.04 |
ENSMUST00000179285.1
|
Hist4h4
|
histone cluster 4, H4 |
| chrX_-_73869804 | 1.03 |
ENSMUST00000066576.5
ENSMUST00000114430.1 |
L1cam
|
L1 cell adhesion molecule |
| chr13_+_21735055 | 1.01 |
ENSMUST00000087714.4
|
Hist1h4j
|
histone cluster 1, H4j |
| chr11_-_70220969 | 1.01 |
ENSMUST00000060010.2
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr9_+_106429537 | 0.98 |
ENSMUST00000059802.6
|
Rpl29
|
ribosomal protein L29 |
| chr2_+_72476159 | 0.96 |
ENSMUST00000102691.4
|
Cdca7
|
cell division cycle associated 7 |
| chr9_+_106429399 | 0.94 |
ENSMUST00000150576.1
|
Rpl29
|
ribosomal protein L29 |
| chr18_-_77713978 | 0.91 |
ENSMUST00000074653.4
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr7_-_44974781 | 0.90 |
ENSMUST00000063761.7
|
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr17_+_28328471 | 0.90 |
ENSMUST00000042334.8
|
Rpl10a
|
ribosomal protein L10A |
| chr2_+_72476225 | 0.88 |
ENSMUST00000157019.1
|
Cdca7
|
cell division cycle associated 7 |
| chr9_+_121719403 | 0.87 |
ENSMUST00000182225.1
|
Nktr
|
natural killer tumor recognition sequence |
| chr9_+_70012540 | 0.80 |
ENSMUST00000118198.1
ENSMUST00000119905.1 ENSMUST00000119413.1 ENSMUST00000140305.1 ENSMUST00000122087.1 |
Gtf2a2
|
general transcription factor II A, 2 |
| chr8_-_11008458 | 0.79 |
ENSMUST00000040514.6
|
Irs2
|
insulin receptor substrate 2 |
| chr10_-_62602261 | 0.79 |
ENSMUST00000045866.7
|
Ddx21
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 |
| chr3_+_122245557 | 0.76 |
ENSMUST00000029769.7
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr17_+_35879770 | 0.71 |
ENSMUST00000025292.8
|
Dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr3_+_122245625 | 0.70 |
ENSMUST00000178826.1
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr4_-_70410422 | 0.69 |
ENSMUST00000144099.1
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr11_-_70410010 | 0.68 |
ENSMUST00000019065.3
ENSMUST00000135148.1 |
Pelp1
|
proline, glutamic acid and leucine rich protein 1 |
| chr13_-_25020289 | 0.60 |
ENSMUST00000021772.2
|
Mrs2
|
MRS2 magnesium homeostasis factor homolog (S. cerevisiae) |
| chr18_-_34651703 | 0.56 |
ENSMUST00000025228.5
ENSMUST00000133181.1 |
Cdc23
|
CDC23 cell division cycle 23 |
| chr7_-_102494773 | 0.55 |
ENSMUST00000058750.3
|
Olfr545
|
olfactory receptor 545 |
| chr5_+_121397936 | 0.55 |
ENSMUST00000042163.8
|
Naa25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr9_+_121719172 | 0.54 |
ENSMUST00000035112.6
ENSMUST00000182311.1 |
Nktr
|
natural killer tumor recognition sequence |
| chr13_+_37825975 | 0.48 |
ENSMUST00000138043.1
|
Rreb1
|
ras responsive element binding protein 1 |
| chrX_-_147554050 | 0.46 |
ENSMUST00000112819.2
ENSMUST00000136789.1 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr13_+_51645232 | 0.45 |
ENSMUST00000075853.5
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr13_+_14063776 | 0.44 |
ENSMUST00000129488.1
ENSMUST00000110536.1 ENSMUST00000110534.1 ENSMUST00000039538.8 ENSMUST00000110533.1 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr4_-_116627478 | 0.43 |
ENSMUST00000081182.4
ENSMUST00000030457.5 |
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr2_-_151741236 | 0.41 |
ENSMUST00000042452.4
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr10_+_79755103 | 0.41 |
ENSMUST00000020577.2
|
Fgf22
|
fibroblast growth factor 22 |
| chr7_-_25816616 | 0.39 |
ENSMUST00000043314.3
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr15_+_40655020 | 0.39 |
ENSMUST00000053467.4
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr11_-_84916338 | 0.31 |
ENSMUST00000103195.4
|
Znhit3
|
zinc finger, HIT type 3 |
| chr8_-_54724474 | 0.30 |
ENSMUST00000175915.1
|
Wdr17
|
WD repeat domain 17 |
| chr3_+_95658771 | 0.28 |
ENSMUST00000178686.1
|
Mcl1
|
myeloid cell leukemia sequence 1 |
| chr7_+_78783119 | 0.22 |
ENSMUST00000032840.4
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr7_+_28808795 | 0.21 |
ENSMUST00000172529.1
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr3_+_95658714 | 0.21 |
ENSMUST00000037947.8
|
Mcl1
|
myeloid cell leukemia sequence 1 |
| chr2_+_155517948 | 0.19 |
ENSMUST00000029135.8
ENSMUST00000065973.2 ENSMUST00000103142.5 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr11_+_101009452 | 0.17 |
ENSMUST00000044721.6
ENSMUST00000103110.3 ENSMUST00000168757.2 |
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr13_-_14063395 | 0.14 |
ENSMUST00000170957.1
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr8_-_27128615 | 0.13 |
ENSMUST00000033877.4
|
Brf2
|
BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like |
| chrX_-_73458990 | 0.13 |
ENSMUST00000033737.8
ENSMUST00000077243.4 |
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr1_-_192137998 | 0.12 |
ENSMUST00000073279.7
ENSMUST00000110849.3 |
Rcor3
|
REST corepressor 3 |
| chr2_-_71546745 | 0.12 |
ENSMUST00000024159.6
|
Dlx2
|
distal-less homeobox 2 |
| chr6_-_100671126 | 0.11 |
ENSMUST00000089245.6
ENSMUST00000113312.2 ENSMUST00000170667.1 |
Shq1
|
SHQ1 homolog (S. cerevisiae) |
| chr7_+_142303502 | 0.11 |
ENSMUST00000061403.4
|
Krtap5-4
|
keratin associated protein 5-4 |
| chr17_-_6961156 | 0.09 |
ENSMUST00000063683.6
|
Tagap1
|
T cell activation GTPase activating protein 1 |
| chr1_+_159232299 | 0.07 |
ENSMUST00000076894.5
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr9_-_20898592 | 0.07 |
ENSMUST00000004206.8
|
Eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr11_-_32267547 | 0.07 |
ENSMUST00000109389.2
ENSMUST00000129010.1 ENSMUST00000020530.5 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr10_+_128083273 | 0.06 |
ENSMUST00000026459.5
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr9_+_110532501 | 0.06 |
ENSMUST00000153838.2
|
Setd2
|
SET domain containing 2 |
| chr4_+_134343181 | 0.06 |
ENSMUST00000105873.1
ENSMUST00000105874.2 |
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr7_-_47982296 | 0.04 |
ENSMUST00000087092.2
|
Mrgpra4
|
MAS-related GPR, member A4 |
| chr1_+_40465976 | 0.00 |
ENSMUST00000108044.2
ENSMUST00000087983.2 |
Il18r1
|
interleukin 18 receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.6 | 13.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.5 | 1.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 1.3 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.4 | 2.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 1.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.2 | 1.5 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.2 | 1.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 3.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.4 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 1.1 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 4.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.0 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.8 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 1.2 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.1 | 0.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.9 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.8 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.8 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 2.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 1.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.4 | 1.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 11.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 4.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.5 | 1.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.5 | 4.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 1.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.4 | 3.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 1.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.4 | 1.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 3.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.9 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 13.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 3.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.7 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 3.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 5.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 11.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 2.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |