Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
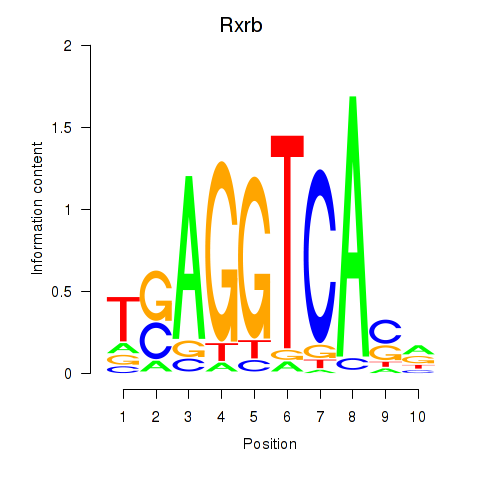
Results for Rxrb
Z-value: 0.49
Transcription factors associated with Rxrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxrb
|
ENSMUSG00000039656.10 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrb | mm10_v2_chr17_+_34031787_34031821 | -0.56 | 4.3e-04 | Click! |
Activity profile of Rxrb motif
Sorted Z-values of Rxrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_52824340 | 2.46 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr2_+_102706356 | 2.17 |
ENSMUST00000123759.1
ENSMUST00000111212.1 ENSMUST00000005220.4 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_+_167618246 | 2.12 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr12_-_113260217 | 1.63 |
ENSMUST00000178282.1
|
Igha
|
immunoglobulin heavy constant alpha |
| chr3_-_89393629 | 1.55 |
ENSMUST00000124783.1
ENSMUST00000126027.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr8_-_67910911 | 1.42 |
ENSMUST00000093468.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_180193653 | 1.42 |
ENSMUST00000159914.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr2_+_72054598 | 1.39 |
ENSMUST00000028525.5
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_33314375 | 1.38 |
ENSMUST00000155386.1
|
Atxn7l1
|
ataxin 7-like 1 |
| chr1_-_140183404 | 1.21 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr15_+_76268076 | 1.17 |
ENSMUST00000074173.3
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr17_+_24878724 | 1.17 |
ENSMUST00000050714.6
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr1_-_140183283 | 1.15 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr14_-_118052235 | 1.12 |
ENSMUST00000022725.2
|
Dct
|
dopachrome tautomerase |
| chr7_-_141434402 | 1.06 |
ENSMUST00000136354.1
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_-_141434532 | 0.98 |
ENSMUST00000133021.1
ENSMUST00000106007.3 ENSMUST00000150026.1 ENSMUST00000133206.2 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr4_+_43632185 | 0.98 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr7_-_105600103 | 0.89 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr6_-_124863877 | 0.88 |
ENSMUST00000046893.7
|
Gpr162
|
G protein-coupled receptor 162 |
| chr5_-_116422858 | 0.86 |
ENSMUST00000036991.4
|
Hspb8
|
heat shock protein 8 |
| chr11_+_75193783 | 0.85 |
ENSMUST00000102514.3
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chr11_-_3722189 | 0.85 |
ENSMUST00000102950.3
ENSMUST00000101632.3 |
Osbp2
|
oxysterol binding protein 2 |
| chr8_+_76899772 | 0.85 |
ENSMUST00000109913.2
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr1_-_157108632 | 0.81 |
ENSMUST00000118207.1
ENSMUST00000027884.6 ENSMUST00000121911.1 |
Tex35
|
testis expressed 35 |
| chrX_-_100412587 | 0.81 |
ENSMUST00000033567.8
|
Awat2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr14_-_16243309 | 0.80 |
ENSMUST00000112625.1
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr11_+_79660532 | 0.76 |
ENSMUST00000155381.1
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr14_-_47189406 | 0.72 |
ENSMUST00000089959.6
|
Gch1
|
GTP cyclohydrolase 1 |
| chr16_+_31663841 | 0.72 |
ENSMUST00000115201.1
|
Dlg1
|
discs, large homolog 1 (Drosophila) |
| chr10_+_93641041 | 0.71 |
ENSMUST00000020204.4
|
Ntn4
|
netrin 4 |
| chr4_+_136143497 | 0.67 |
ENSMUST00000008016.2
|
Id3
|
inhibitor of DNA binding 3 |
| chr6_+_147531392 | 0.66 |
ENSMUST00000111614.2
|
Ccdc91
|
coiled-coil domain containing 91 |
| chr7_-_126922887 | 0.66 |
ENSMUST00000134134.1
ENSMUST00000119781.1 ENSMUST00000121612.2 |
Tmem219
|
transmembrane protein 219 |
| chr16_-_23520579 | 0.64 |
ENSMUST00000089883.5
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr10_+_61720592 | 0.63 |
ENSMUST00000080099.5
|
Aifm2
|
apoptosis-inducing factor, mitochondrion-associated 2 |
| chr11_-_50325599 | 0.62 |
ENSMUST00000179865.1
ENSMUST00000020637.8 |
Canx
|
calnexin |
| chr6_+_129533183 | 0.61 |
ENSMUST00000032264.6
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr14_-_21741476 | 0.59 |
ENSMUST00000184703.1
ENSMUST00000183698.1 ENSMUST00000119866.2 |
Dusp13
|
dual specificity phosphatase 13 |
| chr9_-_106891870 | 0.59 |
ENSMUST00000160503.1
ENSMUST00000159620.2 ENSMUST00000160978.1 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chrX_+_73716712 | 0.57 |
ENSMUST00000114461.2
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr16_+_31663935 | 0.56 |
ENSMUST00000100001.3
ENSMUST00000064477.7 |
Dlg1
|
discs, large homolog 1 (Drosophila) |
| chr6_+_90302083 | 0.56 |
ENSMUST00000167550.2
|
BC048671
|
cDNA sequence BC048671 |
| chr5_+_119834663 | 0.55 |
ENSMUST00000018407.6
|
Tbx5
|
T-box 5 |
| chr12_+_33314277 | 0.52 |
ENSMUST00000133549.1
|
Atxn7l1
|
ataxin 7-like 1 |
| chrX_+_73716577 | 0.50 |
ENSMUST00000002084.7
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chrX_+_101376359 | 0.50 |
ENSMUST00000119080.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr4_-_20778852 | 0.49 |
ENSMUST00000102998.3
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr10_-_17947997 | 0.49 |
ENSMUST00000037879.6
|
Heca
|
headcase homolog (Drosophila) |
| chr16_+_24721842 | 0.48 |
ENSMUST00000115314.2
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_6212979 | 0.48 |
ENSMUST00000114941.1
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr6_-_29380426 | 0.47 |
ENSMUST00000147483.2
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr2_-_6213033 | 0.46 |
ENSMUST00000042658.2
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr5_+_72914554 | 0.45 |
ENSMUST00000143829.1
|
Slain2
|
SLAIN motif family, member 2 |
| chr16_-_30388530 | 0.45 |
ENSMUST00000100013.2
ENSMUST00000061350.6 |
Atp13a3
|
ATPase type 13A3 |
| chr5_+_91139591 | 0.45 |
ENSMUST00000031325.4
|
Areg
|
amphiregulin |
| chr3_-_94786430 | 0.44 |
ENSMUST00000107272.1
|
Cgn
|
cingulin |
| chr8_+_105708270 | 0.44 |
ENSMUST00000013302.5
|
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr9_+_55208925 | 0.44 |
ENSMUST00000034859.8
|
Fbxo22
|
F-box protein 22 |
| chr5_-_74702891 | 0.42 |
ENSMUST00000117388.1
|
Lnx1
|
ligand of numb-protein X 1 |
| chr17_-_27820445 | 0.41 |
ENSMUST00000114859.1
|
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr7_-_31055594 | 0.41 |
ENSMUST00000039909.6
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr16_+_10545390 | 0.41 |
ENSMUST00000115827.1
ENSMUST00000038145.6 ENSMUST00000150894.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr3_+_90062781 | 0.40 |
ENSMUST00000029551.2
|
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr6_-_113531575 | 0.40 |
ENSMUST00000032425.5
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr12_-_98737405 | 0.40 |
ENSMUST00000170188.1
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr17_-_27820534 | 0.40 |
ENSMUST00000075076.4
ENSMUST00000114863.2 |
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chr6_-_124965403 | 0.39 |
ENSMUST00000129446.1
ENSMUST00000032220.8 |
Cops7a
|
COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) |
| chr5_-_123666682 | 0.39 |
ENSMUST00000149410.1
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr11_-_53430779 | 0.37 |
ENSMUST00000061326.4
ENSMUST00000109021.3 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr11_+_40733639 | 0.37 |
ENSMUST00000020578.4
|
Nudcd2
|
NudC domain containing 2 |
| chr5_-_24995748 | 0.36 |
ENSMUST00000076306.5
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_+_101070012 | 0.36 |
ENSMUST00000001802.9
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr12_-_111712946 | 0.36 |
ENSMUST00000160825.1
ENSMUST00000162953.1 |
Bag5
|
BCL2-associated athanogene 5 |
| chr1_+_84839833 | 0.36 |
ENSMUST00000097672.3
|
Fbxo36
|
F-box protein 36 |
| chr3_-_94786469 | 0.35 |
ENSMUST00000107273.1
|
Cgn
|
cingulin |
| chr19_+_42052228 | 0.35 |
ENSMUST00000164518.2
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr14_+_21500879 | 0.34 |
ENSMUST00000182964.1
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr16_+_10545339 | 0.34 |
ENSMUST00000066345.7
ENSMUST00000115824.3 ENSMUST00000155633.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr10_-_128821576 | 0.34 |
ENSMUST00000026409.3
|
Ormdl2
|
ORM1-like 2 (S. cerevisiae) |
| chr6_-_124965248 | 0.34 |
ENSMUST00000129976.1
|
Cops7a
|
COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) |
| chr11_-_97629685 | 0.34 |
ENSMUST00000052281.4
|
E130012A19Rik
|
RIKEN cDNA E130012A19 gene |
| chr6_-_124965207 | 0.34 |
ENSMUST00000148485.1
|
Cops7a
|
COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) |
| chr6_+_22288221 | 0.33 |
ENSMUST00000128245.1
ENSMUST00000031681.3 ENSMUST00000148639.1 |
Wnt16
|
wingless-related MMTV integration site 16 |
| chr13_+_46669517 | 0.33 |
ENSMUST00000099547.3
|
C78339
|
expressed sequence C78339 |
| chr5_+_121795034 | 0.32 |
ENSMUST00000162327.1
|
Atxn2
|
ataxin 2 |
| chr1_-_191164815 | 0.32 |
ENSMUST00000171798.1
|
Fam71a
|
family with sequence similarity 71, member A |
| chr15_-_82354280 | 0.32 |
ENSMUST00000023085.5
|
Ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (B14) |
| chr2_-_122313644 | 0.32 |
ENSMUST00000147788.1
ENSMUST00000154412.1 ENSMUST00000110537.1 ENSMUST00000148417.1 |
Duoxa1
|
dual oxidase maturation factor 1 |
| chr9_-_50659780 | 0.32 |
ENSMUST00000034567.3
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr19_-_37176055 | 0.31 |
ENSMUST00000142973.1
ENSMUST00000154376.1 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_+_119945957 | 0.31 |
ENSMUST00000058644.8
|
Tmem196
|
transmembrane protein 196 |
| chr11_-_99620432 | 0.31 |
ENSMUST00000073853.2
|
Gm11562
|
predicted gene 11562 |
| chr17_+_28177339 | 0.30 |
ENSMUST00000073534.2
ENSMUST00000002318.1 |
Zfp523
|
zinc finger protein 523 |
| chr6_-_88446491 | 0.30 |
ENSMUST00000165242.1
|
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr15_-_101562889 | 0.29 |
ENSMUST00000023714.4
|
4732456N10Rik
|
RIKEN cDNA 4732456N10 gene |
| chr7_+_45514581 | 0.29 |
ENSMUST00000151506.1
ENSMUST00000085331.5 ENSMUST00000126061.1 |
Tulp2
|
tubby-like protein 2 |
| chr6_-_29380513 | 0.29 |
ENSMUST00000080428.6
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr19_+_11770415 | 0.28 |
ENSMUST00000167199.1
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr11_+_40733936 | 0.27 |
ENSMUST00000127382.1
|
Nudcd2
|
NudC domain containing 2 |
| chr7_-_120095177 | 0.27 |
ENSMUST00000046993.3
|
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr12_+_108792946 | 0.27 |
ENSMUST00000021692.7
|
Yy1
|
YY1 transcription factor |
| chr7_+_138846579 | 0.27 |
ENSMUST00000155672.1
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta isoform |
| chr4_-_139968026 | 0.26 |
ENSMUST00000105031.2
|
Klhdc7a
|
kelch domain containing 7A |
| chr7_-_35802968 | 0.25 |
ENSMUST00000061586.4
|
Zfp507
|
zinc finger protein 507 |
| chr6_-_124965485 | 0.25 |
ENSMUST00000112439.2
|
Cops7a
|
COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) |
| chr18_-_80713062 | 0.25 |
ENSMUST00000170905.1
ENSMUST00000078049.4 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr4_+_57845240 | 0.24 |
ENSMUST00000102903.1
ENSMUST00000107598.3 |
Akap2
|
A kinase (PRKA) anchor protein 2 |
| chr5_+_72914264 | 0.24 |
ENSMUST00000144843.1
|
Slain2
|
SLAIN motif family, member 2 |
| chr2_+_173659834 | 0.24 |
ENSMUST00000109110.3
|
Rab22a
|
RAB22A, member RAS oncogene family |
| chr11_-_106973090 | 0.23 |
ENSMUST00000150366.1
|
Gm11707
|
predicted gene 11707 |
| chr15_+_38219203 | 0.23 |
ENSMUST00000081966.4
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr12_-_111712909 | 0.23 |
ENSMUST00000160576.1
|
Bag5
|
BCL2-associated athanogene 5 |
| chr7_-_4778141 | 0.22 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr17_-_67632707 | 0.21 |
ENSMUST00000097290.2
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr1_+_160044374 | 0.21 |
ENSMUST00000163892.1
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chr4_-_58912678 | 0.21 |
ENSMUST00000144512.1
ENSMUST00000102889.3 ENSMUST00000055822.8 |
AI314180
|
expressed sequence AI314180 |
| chr19_-_6980420 | 0.21 |
ENSMUST00000070878.8
ENSMUST00000177752.1 |
Fkbp2
|
FK506 binding protein 2 |
| chr1_+_6487231 | 0.21 |
ENSMUST00000140079.1
ENSMUST00000131494.1 |
St18
|
suppression of tumorigenicity 18 |
| chr7_+_102229999 | 0.20 |
ENSMUST00000120119.1
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr16_-_10790902 | 0.20 |
ENSMUST00000050864.5
|
Prm3
|
protamine 3 |
| chr7_-_4771262 | 0.20 |
ENSMUST00000108580.2
|
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr16_-_44333135 | 0.20 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr5_+_3803160 | 0.19 |
ENSMUST00000171023.1
ENSMUST00000080085.4 |
Krit1
|
KRIT1, ankyrin repeat containing |
| chr3_-_89773221 | 0.19 |
ENSMUST00000038450.1
|
4632404H12Rik
|
RIKEN cDNA 4632404H12 gene |
| chr7_-_4771279 | 0.19 |
ENSMUST00000174409.1
|
Fam71e2
|
family with sequence similarity 71, member E2 |
| chr1_-_9631092 | 0.18 |
ENSMUST00000115480.1
|
2610203C22Rik
|
RIKEN cDNA 2610203C22 gene |
| chr4_-_45532470 | 0.17 |
ENSMUST00000147448.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr7_+_138846335 | 0.17 |
ENSMUST00000041097.6
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta isoform |
| chr6_+_96115249 | 0.17 |
ENSMUST00000075080.5
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr12_-_111713185 | 0.17 |
ENSMUST00000054636.6
|
Bag5
|
BCL2-associated athanogene 5 |
| chr11_+_104186327 | 0.17 |
ENSMUST00000107000.1
ENSMUST00000059448.7 |
Sppl2c
|
signal peptide peptidase 2C |
| chr1_+_160044564 | 0.17 |
ENSMUST00000168359.1
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chr2_-_119662756 | 0.16 |
ENSMUST00000028768.1
ENSMUST00000110801.1 ENSMUST00000110802.1 |
Ndufaf1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 |
| chr4_-_20778527 | 0.16 |
ENSMUST00000119374.1
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr5_+_25516061 | 0.16 |
ENSMUST00000045016.7
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr1_+_131688766 | 0.16 |
ENSMUST00000129905.1
|
5430435G22Rik
|
RIKEN cDNA 5430435G22 gene |
| chr4_+_150997081 | 0.16 |
ENSMUST00000030803.1
|
Uts2
|
urotensin 2 |
| chr12_+_86241848 | 0.16 |
ENSMUST00000071106.4
|
Gpatch2l
|
G patch domain containing 2 like |
| chr12_+_111713255 | 0.15 |
ENSMUST00000040519.5
ENSMUST00000160366.1 ENSMUST00000163220.2 ENSMUST00000162316.1 |
Apopt1
|
apoptogenic, mitochondrial 1 |
| chr13_-_54688246 | 0.15 |
ENSMUST00000122935.1
ENSMUST00000128257.1 |
Rnf44
|
ring finger protein 44 |
| chr7_-_104129792 | 0.14 |
ENSMUST00000053743.4
|
4931431F19Rik
|
RIKEN cDNA 4931431F19 gene |
| chr2_-_151092350 | 0.14 |
ENSMUST00000109890.1
|
Gm14151
|
predicted gene 14151 |
| chr13_+_55209776 | 0.14 |
ENSMUST00000099490.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr1_-_171531660 | 0.13 |
ENSMUST00000168184.1
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chrX_-_23365044 | 0.13 |
ENSMUST00000115313.1
|
Klhl13
|
kelch-like 13 |
| chr3_+_98160812 | 0.13 |
ENSMUST00000050342.3
|
Adam30
|
a disintegrin and metallopeptidase domain 30 |
| chr17_+_28575718 | 0.13 |
ENSMUST00000080780.6
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr11_+_117484368 | 0.13 |
ENSMUST00000092394.3
|
Gm11733
|
predicted gene 11733 |
| chr8_+_109493982 | 0.12 |
ENSMUST00000034162.6
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chrX_+_86191764 | 0.12 |
ENSMUST00000026036.4
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr6_+_90269106 | 0.12 |
ENSMUST00000058039.2
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr13_-_55415166 | 0.11 |
ENSMUST00000054146.3
|
Pfn3
|
profilin 3 |
| chr2_-_6721606 | 0.11 |
ENSMUST00000150624.2
ENSMUST00000142941.1 ENSMUST00000100429.4 ENSMUST00000182879.1 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr16_-_44332925 | 0.11 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr9_-_77251871 | 0.11 |
ENSMUST00000183955.1
|
Mlip
|
muscular LMNA-interacting protein |
| chr2_+_173659760 | 0.10 |
ENSMUST00000029024.3
|
Rab22a
|
RAB22A, member RAS oncogene family |
| chr18_+_57142782 | 0.10 |
ENSMUST00000139892.1
|
Megf10
|
multiple EGF-like-domains 10 |
| chr13_-_54688264 | 0.10 |
ENSMUST00000150626.1
ENSMUST00000134177.1 |
Rnf44
|
ring finger protein 44 |
| chr2_+_121449362 | 0.10 |
ENSMUST00000110615.1
ENSMUST00000099475.5 |
Serf2
|
small EDRK-rich factor 2 |
| chr1_+_86022105 | 0.10 |
ENSMUST00000131151.2
|
Spata3
|
spermatogenesis associated 3 |
| chr4_+_42969945 | 0.09 |
ENSMUST00000030163.5
|
1700022I11Rik
|
RIKEN cDNA 1700022I11 gene |
| chr3_+_94837533 | 0.09 |
ENSMUST00000107270.2
|
Pogz
|
pogo transposable element with ZNF domain |
| chr6_+_71543900 | 0.08 |
ENSMUST00000065364.2
|
Chmp3
|
charged multivesicular body protein 3 |
| chr10_+_80148263 | 0.08 |
ENSMUST00000099492.3
ENSMUST00000042057.5 |
Midn
|
midnolin |
| chr2_+_118900377 | 0.08 |
ENSMUST00000151162.1
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr2_-_119477613 | 0.08 |
ENSMUST00000110808.1
ENSMUST00000049920.7 |
Ino80
|
INO80 homolog (S. cerevisiae) |
| chr15_+_92161343 | 0.08 |
ENSMUST00000068378.5
|
Cntn1
|
contactin 1 |
| chr4_+_148000722 | 0.07 |
ENSMUST00000103230.4
|
Nppa
|
natriuretic peptide type A |
| chr6_+_71543797 | 0.07 |
ENSMUST00000059462.5
|
Chmp3
|
charged multivesicular body protein 3 |
| chr11_-_95699143 | 0.07 |
ENSMUST00000062249.2
|
Gm9796
|
predicted gene 9796 |
| chr2_-_153871076 | 0.07 |
ENSMUST00000028982.4
|
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr1_+_175698586 | 0.07 |
ENSMUST00000171939.1
ENSMUST00000094288.4 |
Wdr64
|
WD repeat domain 64 |
| chr9_-_77251829 | 0.05 |
ENSMUST00000184322.1
ENSMUST00000184316.1 |
Mlip
|
muscular LMNA-interacting protein |
| chr6_+_119236507 | 0.04 |
ENSMUST00000037434.6
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr1_+_86021959 | 0.04 |
ENSMUST00000159876.1
ENSMUST00000135440.1 |
Spata3
|
spermatogenesis associated 3 |
| chr17_-_75551838 | 0.04 |
ENSMUST00000112507.3
|
Fam98a
|
family with sequence similarity 98, member A |
| chr10_+_81136223 | 0.04 |
ENSMUST00000048128.8
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr7_+_19212521 | 0.03 |
ENSMUST00000060225.4
|
Gpr4
|
G protein-coupled receptor 4 |
| chr15_+_78430086 | 0.02 |
ENSMUST00000162808.1
|
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chrX_-_112406779 | 0.02 |
ENSMUST00000026601.2
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr11_+_97315716 | 0.01 |
ENSMUST00000019026.3
ENSMUST00000132168.1 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr7_-_134364859 | 0.01 |
ENSMUST00000172947.1
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chrX_+_99042581 | 0.01 |
ENSMUST00000036606.7
|
Stard8
|
START domain containing 8 |
| chr1_+_131688670 | 0.00 |
ENSMUST00000064679.2
ENSMUST00000064664.3 ENSMUST00000136247.1 |
5430435G22Rik
|
RIKEN cDNA 5430435G22 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.4 | 1.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.4 | 1.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 1.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 2.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 0.8 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.3 | 2.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.6 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 0.5 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 0.7 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.2 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.8 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.8 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 1.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.4 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 | 0.3 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.8 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 2.0 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.1 | 0.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.1 | 0.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.4 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 2.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 1.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.3 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.4 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.7 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.8 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 1.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0072144 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 0.7 | GO:0036019 | endolysosome(GO:0036019) |
| 0.2 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 1.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 3.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 2.4 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.3 | 2.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 0.8 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 0.8 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 2.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.8 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.6 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.4 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |