Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
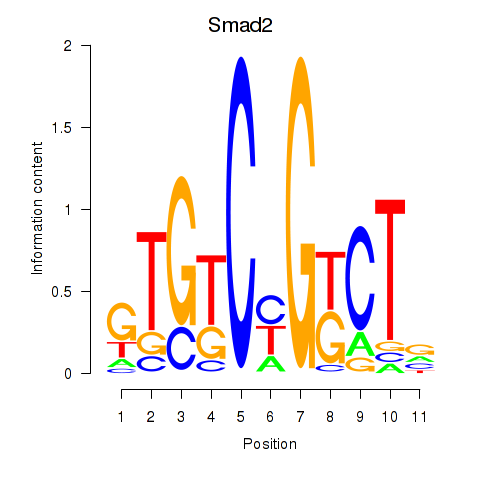
Results for Smad2
Z-value: 0.34
Transcription factors associated with Smad2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad2
|
ENSMUSG00000024563.9 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad2 | mm10_v2_chr18_+_76241892_76242046 | -0.14 | 4.1e-01 | Click! |
Activity profile of Smad2 motif
Sorted Z-values of Smad2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126463100 | 2.08 |
ENSMUST00000032974.6
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr7_+_19411086 | 1.78 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr15_-_101924725 | 1.11 |
ENSMUST00000023797.6
|
Krt4
|
keratin 4 |
| chr6_+_30639218 | 0.79 |
ENSMUST00000031806.9
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr6_-_55175019 | 0.51 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr2_-_84822546 | 0.42 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr15_-_84123174 | 0.39 |
ENSMUST00000019012.3
|
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr7_+_26307190 | 0.38 |
ENSMUST00000098657.3
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr1_-_172297989 | 0.30 |
ENSMUST00000085913.4
ENSMUST00000097464.2 ENSMUST00000137679.1 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr10_-_118868903 | 0.26 |
ENSMUST00000004281.8
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr14_-_20496780 | 0.23 |
ENSMUST00000022353.3
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr7_-_34654342 | 0.22 |
ENSMUST00000108069.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr10_-_49788743 | 0.20 |
ENSMUST00000105483.1
ENSMUST00000105487.1 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr8_-_105637403 | 0.20 |
ENSMUST00000182046.1
|
Gm5914
|
predicted gene 5914 |
| chr3_+_89136133 | 0.17 |
ENSMUST00000047111.6
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr11_-_116654245 | 0.17 |
ENSMUST00000021166.5
|
Cygb
|
cytoglobin |
| chr14_-_12345847 | 0.16 |
ENSMUST00000022262.4
|
Fezf2
|
Fez family zinc finger 2 |
| chr10_+_80150448 | 0.16 |
ENSMUST00000153477.1
|
Midn
|
midnolin |
| chr3_+_89136572 | 0.16 |
ENSMUST00000107482.3
ENSMUST00000127058.1 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr11_+_4135233 | 0.13 |
ENSMUST00000124670.1
|
Rnf215
|
ring finger protein 215 |
| chr2_+_177768044 | 0.13 |
ENSMUST00000108942.3
|
Gm14322
|
predicted gene 14322 |
| chr15_+_57694651 | 0.12 |
ENSMUST00000096430.4
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr17_-_46032366 | 0.12 |
ENSMUST00000071648.5
ENSMUST00000142351.2 ENSMUST00000167860.1 |
Vegfa
|
vascular endothelial growth factor A |
| chr11_-_69122589 | 0.11 |
ENSMUST00000180487.1
|
9130213A22Rik
|
RIKEN cDNA 9130213A22 gene |
| chr15_-_101562889 | 0.11 |
ENSMUST00000023714.4
|
4732456N10Rik
|
RIKEN cDNA 4732456N10 gene |
| chr11_-_33147400 | 0.11 |
ENSMUST00000020507.7
|
Fgf18
|
fibroblast growth factor 18 |
| chr8_-_87959560 | 0.11 |
ENSMUST00000109655.2
|
Zfp423
|
zinc finger protein 423 |
| chr15_+_102102926 | 0.11 |
ENSMUST00000169627.1
ENSMUST00000046144.9 |
Tenc1
|
tensin like C1 domain-containing phosphatase |
| chr10_+_80300997 | 0.10 |
ENSMUST00000140828.1
ENSMUST00000138909.1 |
Apc2
|
adenomatosis polyposis coli 2 |
| chr17_+_69439326 | 0.10 |
ENSMUST00000169935.1
|
A330050F15Rik
|
RIKEN cDNA A330050F15 gene |
| chr17_+_3397189 | 0.09 |
ENSMUST00000072156.6
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr4_+_148591482 | 0.09 |
ENSMUST00000006611.8
|
Srm
|
spermidine synthase |
| chr3_+_116968267 | 0.09 |
ENSMUST00000117592.1
|
4930455H04Rik
|
RIKEN cDNA 4930455H04 gene |
| chr3_+_135438280 | 0.08 |
ENSMUST00000106291.3
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr15_+_82252397 | 0.08 |
ENSMUST00000136948.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr13_-_100104064 | 0.08 |
ENSMUST00000038104.5
|
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr9_+_44107286 | 0.08 |
ENSMUST00000152956.1
ENSMUST00000114815.1 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chrX_-_94123087 | 0.08 |
ENSMUST00000113925.1
|
Zfx
|
zinc finger protein X-linked |
| chr17_-_35980197 | 0.08 |
ENSMUST00000055454.7
|
Prr3
|
proline-rich polypeptide 3 |
| chrX_-_92599572 | 0.08 |
ENSMUST00000113955.1
|
Mageb18
|
melanoma antigen family B, 18 |
| chr8_+_47822143 | 0.08 |
ENSMUST00000079639.2
|
Cldn24
|
claudin 24 |
| chr11_-_75190458 | 0.07 |
ENSMUST00000044949.4
|
Dph1
|
DPH1 homolog (S. cerevisiae) |
| chr13_+_96542727 | 0.07 |
ENSMUST00000077672.4
ENSMUST00000109444.2 |
Col4a3bp
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr3_+_135438722 | 0.07 |
ENSMUST00000166033.1
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_100762928 | 0.07 |
ENSMUST00000107360.2
ENSMUST00000055083.3 |
Hcrt
|
hypocretin |
| chr10_+_5069192 | 0.07 |
ENSMUST00000180100.1
|
Gm10097
|
predicted gene 10097 |
| chr14_-_18331855 | 0.06 |
ENSMUST00000022296.6
|
Ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr17_-_34615776 | 0.06 |
ENSMUST00000168353.2
|
Egfl8
|
EGF-like domain 8 |
| chr13_-_104816908 | 0.06 |
ENSMUST00000022228.6
|
Cwc27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr14_+_21500879 | 0.05 |
ENSMUST00000182964.1
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr16_+_64851991 | 0.05 |
ENSMUST00000067744.7
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr7_+_101321703 | 0.05 |
ENSMUST00000174291.1
ENSMUST00000167888.2 ENSMUST00000172662.1 ENSMUST00000173270.1 ENSMUST00000174083.1 |
Stard10
|
START domain containing 10 |
| chr2_-_128967725 | 0.04 |
ENSMUST00000099385.2
|
Gm10762
|
predicted gene 10762 |
| chr10_-_42583628 | 0.04 |
ENSMUST00000019938.4
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr4_+_42950369 | 0.04 |
ENSMUST00000084662.5
|
Dnajb5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr18_-_42579652 | 0.04 |
ENSMUST00000054738.3
|
Gpr151
|
G protein-coupled receptor 151 |
| chr12_+_33147754 | 0.04 |
ENSMUST00000146040.1
ENSMUST00000125192.1 |
Atxn7l1
|
ataxin 7-like 1 |
| chr3_+_58576636 | 0.04 |
ENSMUST00000107924.1
|
Selt
|
selenoprotein T |
| chr7_-_29180699 | 0.04 |
ENSMUST00000059642.10
|
Psmd8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr6_+_125009261 | 0.04 |
ENSMUST00000112427.1
|
Zfp384
|
zinc finger protein 384 |
| chr7_-_29180454 | 0.03 |
ENSMUST00000182328.1
|
Psmd8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr3_-_83129246 | 0.03 |
ENSMUST00000047876.6
|
Gm10710
|
predicted gene 10710 |
| chr6_+_125009232 | 0.03 |
ENSMUST00000112428.1
|
Zfp384
|
zinc finger protein 384 |
| chr15_+_62178175 | 0.03 |
ENSMUST00000182476.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chrX_-_94123359 | 0.03 |
ENSMUST00000137853.1
ENSMUST00000088102.5 ENSMUST00000113927.1 |
Zfx
|
zinc finger protein X-linked |
| chr3_+_127633134 | 0.03 |
ENSMUST00000029587.7
|
Neurog2
|
neurogenin 2 |
| chr2_+_156863110 | 0.03 |
ENSMUST00000029165.3
|
1110008F13Rik
|
RIKEN cDNA 1110008F13 gene |
| chr10_+_80151154 | 0.02 |
ENSMUST00000146516.1
ENSMUST00000144526.1 |
Midn
|
midnolin |
| chr6_+_125009665 | 0.02 |
ENSMUST00000046064.10
ENSMUST00000152752.1 ENSMUST00000088308.3 ENSMUST00000112425.1 ENSMUST00000084275.5 |
Zfp384
|
zinc finger protein 384 |
| chr11_-_101119814 | 0.02 |
ENSMUST00000107295.3
|
Fam134c
|
family with sequence similarity 134, member C |
| chrX_-_136741155 | 0.02 |
ENSMUST00000166930.1
ENSMUST00000113095.1 ENSMUST00000155207.1 ENSMUST00000080411.6 ENSMUST00000169418.1 |
Morf4l2
|
mortality factor 4 like 2 |
| chr19_+_5878622 | 0.02 |
ENSMUST00000136833.1
ENSMUST00000141362.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr8_-_70439557 | 0.02 |
ENSMUST00000076615.5
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chrX_+_99821021 | 0.02 |
ENSMUST00000096363.2
|
Tmem28
|
transmembrane protein 28 |
| chr11_+_101119938 | 0.02 |
ENSMUST00000043680.8
|
Tubg1
|
tubulin, gamma 1 |
| chr18_+_37307445 | 0.01 |
ENSMUST00000056712.2
|
Pcdhb4
|
protocadherin beta 4 |
| chr18_+_40258361 | 0.01 |
ENSMUST00000091927.4
|
Kctd16
|
potassium channel tetramerisation domain containing 16 |
| chr18_-_31609893 | 0.01 |
ENSMUST00000060396.6
|
Slc25a46
|
solute carrier family 25, member 46 |
| chr11_-_101119889 | 0.01 |
ENSMUST00000017946.5
|
Fam134c
|
family with sequence similarity 134, member C |
| chr14_-_31168587 | 0.01 |
ENSMUST00000036618.7
|
Stab1
|
stabilin 1 |
| chr17_-_34615965 | 0.01 |
ENSMUST00000097345.3
ENSMUST00000015611.7 |
Egfl8
|
EGF-like domain 8 |
| chr18_+_77375065 | 0.01 |
ENSMUST00000123410.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr13_+_23752267 | 0.01 |
ENSMUST00000091703.2
|
Hist1h3b
|
histone cluster 1, H3b |
| chr12_+_33147693 | 0.01 |
ENSMUST00000077456.6
ENSMUST00000110824.2 |
Atxn7l1
|
ataxin 7-like 1 |
| chr8_-_69184177 | 0.01 |
ENSMUST00000185176.1
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr6_-_6882068 | 0.01 |
ENSMUST00000142635.1
ENSMUST00000052609.8 |
Dlx5
|
distal-less homeobox 5 |
| chr10_-_5069044 | 0.00 |
ENSMUST00000095899.3
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0021853 | forebrain anterior/posterior pattern specification(GO:0021797) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 1.8 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |