Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
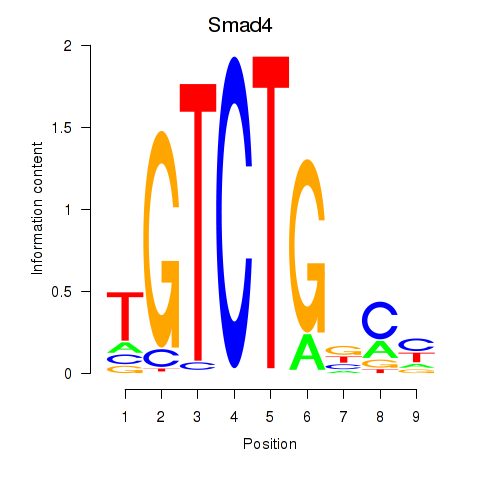
Results for Smad4
Z-value: 0.80
Transcription factors associated with Smad4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad4
|
ENSMUSG00000024515.7 | SMAD family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad4 | mm10_v2_chr18_-_73703739_73703806 | 0.03 | 8.8e-01 | Click! |
Activity profile of Smad4 motif
Sorted Z-values of Smad4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_137288273 | 4.17 |
ENSMUST00000024099.4
ENSMUST00000085934.3 |
Ache
|
acetylcholinesterase |
| chrX_+_136270253 | 3.85 |
ENSMUST00000178632.1
ENSMUST00000053540.4 |
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr4_-_119189949 | 3.58 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chrX_+_136270302 | 3.12 |
ENSMUST00000113112.1
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr4_-_134254076 | 2.65 |
ENSMUST00000060050.5
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr11_-_33147400 | 2.46 |
ENSMUST00000020507.7
|
Fgf18
|
fibroblast growth factor 18 |
| chr4_-_119190005 | 2.36 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr7_+_45216671 | 2.33 |
ENSMUST00000134420.1
|
Tead2
|
TEA domain family member 2 |
| chr4_-_117178726 | 2.29 |
ENSMUST00000153953.1
ENSMUST00000106436.1 |
Kif2c
|
kinesin family member 2C |
| chr17_-_32403551 | 2.20 |
ENSMUST00000135618.1
ENSMUST00000063824.7 |
Rasal3
|
RAS protein activator like 3 |
| chr7_+_45215753 | 1.95 |
ENSMUST00000033060.6
ENSMUST00000155313.1 ENSMUST00000107801.1 |
Tead2
|
TEA domain family member 2 |
| chr7_+_24862193 | 1.89 |
ENSMUST00000052897.4
ENSMUST00000170837.2 |
Gm9844
Gm9844
|
predicted pseudogene 9844 predicted pseudogene 9844 |
| chr7_-_100855403 | 1.85 |
ENSMUST00000156855.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr14_-_57826128 | 1.84 |
ENSMUST00000022536.2
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr17_+_32403006 | 1.83 |
ENSMUST00000065921.5
|
A530088E08Rik
|
RIKEN cDNA A530088E08 gene |
| chr17_-_32403526 | 1.77 |
ENSMUST00000137458.1
|
Rasal3
|
RAS protein activator like 3 |
| chr7_+_117380937 | 1.74 |
ENSMUST00000032892.5
|
Xylt1
|
xylosyltransferase 1 |
| chr15_-_103252810 | 1.72 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr11_-_3504766 | 1.70 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr15_+_39076885 | 1.68 |
ENSMUST00000067072.3
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr12_-_109600328 | 1.54 |
ENSMUST00000149046.2
|
Rtl1
|
retrotransposon-like 1 |
| chr1_-_45503282 | 1.50 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chr13_-_23551648 | 1.46 |
ENSMUST00000102971.1
|
Hist1h4f
|
histone cluster 1, H4f |
| chr17_+_34597852 | 1.42 |
ENSMUST00000174496.2
ENSMUST00000015596.3 ENSMUST00000173992.1 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr2_+_120476911 | 1.39 |
ENSMUST00000110716.1
ENSMUST00000028748.6 ENSMUST00000090028.5 ENSMUST00000110719.2 |
Capn3
|
calpain 3 |
| chr2_+_29869484 | 1.35 |
ENSMUST00000047521.6
ENSMUST00000134152.1 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr1_+_87470258 | 1.33 |
ENSMUST00000027476.4
|
3110079O15Rik
|
RIKEN cDNA 3110079O15 gene |
| chr13_-_22042949 | 1.25 |
ENSMUST00000091741.4
|
Hist1h2ag
|
histone cluster 1, H2ag |
| chr17_-_29888570 | 1.24 |
ENSMUST00000171691.1
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr13_+_22043189 | 1.22 |
ENSMUST00000110452.1
|
Hist1h2bj
|
histone cluster 1, H2bj |
| chr6_+_147032528 | 1.21 |
ENSMUST00000036194.4
|
Rep15
|
RAB15 effector protein |
| chr11_-_106160101 | 1.21 |
ENSMUST00000045923.3
|
Limd2
|
LIM domain containing 2 |
| chr14_+_54640952 | 1.20 |
ENSMUST00000169818.2
|
Gm17606
|
predicted gene, 17606 |
| chr15_-_27681498 | 1.17 |
ENSMUST00000100739.3
|
Fam105a
|
family with sequence similarity 105, member A |
| chr9_-_45936049 | 1.15 |
ENSMUST00000034590.2
|
Tagln
|
transgelin |
| chr5_-_137741102 | 1.12 |
ENSMUST00000149512.1
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr4_+_130055010 | 1.12 |
ENSMUST00000123617.1
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr17_-_28486082 | 1.10 |
ENSMUST00000079413.3
|
Fkbp5
|
FK506 binding protein 5 |
| chr9_-_49798905 | 1.09 |
ENSMUST00000114476.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr18_-_52529847 | 1.08 |
ENSMUST00000171470.1
|
Lox
|
lysyl oxidase |
| chr9_-_14381242 | 1.07 |
ENSMUST00000167549.1
|
Endod1
|
endonuclease domain containing 1 |
| chr15_+_81936753 | 1.04 |
ENSMUST00000038757.7
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr6_-_72958097 | 1.03 |
ENSMUST00000114049.1
|
Tmsb10
|
thymosin, beta 10 |
| chr8_-_92356103 | 1.01 |
ENSMUST00000034183.3
|
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr6_+_120093348 | 1.00 |
ENSMUST00000112711.2
|
Ninj2
|
ninjurin 2 |
| chr18_-_52529692 | 1.00 |
ENSMUST00000025409.7
|
Lox
|
lysyl oxidase |
| chr7_-_27181149 | 0.99 |
ENSMUST00000071986.6
ENSMUST00000121848.1 |
Mia
|
melanoma inhibitory activity |
| chr2_-_29869785 | 0.99 |
ENSMUST00000047607.1
|
2600006K01Rik
|
RIKEN cDNA 2600006K01 gene |
| chr11_-_26591729 | 0.99 |
ENSMUST00000109504.1
|
Vrk2
|
vaccinia related kinase 2 |
| chrX_-_150812932 | 0.96 |
ENSMUST00000131241.1
ENSMUST00000147152.1 ENSMUST00000143843.1 |
Maged2
|
melanoma antigen, family D, 2 |
| chr6_-_72958465 | 0.96 |
ENSMUST00000114050.1
|
Tmsb10
|
thymosin, beta 10 |
| chr7_-_45052865 | 0.91 |
ENSMUST00000057293.6
|
Prr12
|
proline rich 12 |
| chr11_-_106159902 | 0.89 |
ENSMUST00000064545.4
|
Limd2
|
LIM domain containing 2 |
| chr7_-_45870928 | 0.89 |
ENSMUST00000146672.1
|
Grin2d
|
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
| chr7_-_45239041 | 0.88 |
ENSMUST00000131290.1
|
Cd37
|
CD37 antigen |
| chr15_+_81936911 | 0.88 |
ENSMUST00000135663.1
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr3_+_88607742 | 0.87 |
ENSMUST00000175903.1
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr17_+_75178911 | 0.86 |
ENSMUST00000112514.1
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr17_+_75178797 | 0.86 |
ENSMUST00000112516.1
ENSMUST00000135447.1 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chrX_+_10485121 | 0.86 |
ENSMUST00000076354.6
ENSMUST00000115526.1 |
Tspan7
|
tetraspanin 7 |
| chr1_-_133801031 | 0.85 |
ENSMUST00000143567.1
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_-_106160708 | 0.85 |
ENSMUST00000106875.1
|
Limd2
|
LIM domain containing 2 |
| chr9_-_49798729 | 0.84 |
ENSMUST00000166811.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr11_+_69846610 | 0.83 |
ENSMUST00000152566.1
ENSMUST00000108633.2 |
Plscr3
|
phospholipid scramblase 3 |
| chr6_-_129275360 | 0.83 |
ENSMUST00000032259.3
|
Cd69
|
CD69 antigen |
| chr7_-_45238794 | 0.82 |
ENSMUST00000098461.1
ENSMUST00000107797.1 |
Cd37
|
CD37 antigen |
| chr13_-_21832194 | 0.80 |
ENSMUST00000102979.1
|
Hist1h4n
|
histone cluster 1, H4n |
| chrX_+_159708593 | 0.79 |
ENSMUST00000080394.6
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_+_69846665 | 0.78 |
ENSMUST00000019605.2
|
Plscr3
|
phospholipid scramblase 3 |
| chr3_-_102964124 | 0.77 |
ENSMUST00000058899.8
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr11_+_59542748 | 0.76 |
ENSMUST00000149126.1
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chrX_+_56779437 | 0.76 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr10_-_91082704 | 0.74 |
ENSMUST00000162618.1
ENSMUST00000020157.6 ENSMUST00000160788.1 |
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr11_-_95514570 | 0.73 |
ENSMUST00000058866.7
|
Nxph3
|
neurexophilin 3 |
| chr10_-_91082653 | 0.73 |
ENSMUST00000159110.1
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr15_-_98728120 | 0.72 |
ENSMUST00000003445.6
|
Fkbp11
|
FK506 binding protein 11 |
| chr11_+_35121126 | 0.72 |
ENSMUST00000069837.3
|
Slit3
|
slit homolog 3 (Drosophila) |
| chr19_-_31664356 | 0.72 |
ENSMUST00000073581.5
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr4_-_148130678 | 0.71 |
ENSMUST00000030862.4
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr5_+_19907502 | 0.71 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_+_68572245 | 0.69 |
ENSMUST00000170788.2
|
Schip1
|
schwannomin interacting protein 1 |
| chr4_-_115781012 | 0.69 |
ENSMUST00000106521.1
|
Tex38
|
testis expressed 38 |
| chr12_-_100899436 | 0.68 |
ENSMUST00000053668.3
|
Gpr68
|
G protein-coupled receptor 68 |
| chr16_-_36642742 | 0.67 |
ENSMUST00000135280.1
|
Cd86
|
CD86 antigen |
| chr15_-_98778150 | 0.67 |
ENSMUST00000023732.5
|
Wnt10b
|
wingless related MMTV integration site 10b |
| chr14_+_53683593 | 0.66 |
ENSMUST00000103663.4
|
Trav4-4-dv10
|
T cell receptor alpha variable 4-4-DV10 |
| chr8_+_57332111 | 0.65 |
ENSMUST00000181638.1
|
5033428I22Rik
|
RIKEN cDNA 5033428I22 gene |
| chr11_+_60699718 | 0.64 |
ENSMUST00000052346.3
|
Llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr9_+_53771499 | 0.61 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr4_+_127172866 | 0.58 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr11_+_60699758 | 0.58 |
ENSMUST00000108719.3
|
Llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr5_-_92278155 | 0.54 |
ENSMUST00000159345.1
ENSMUST00000113102.3 |
Naaa
|
N-acylethanolamine acid amidase |
| chr4_-_148500449 | 0.54 |
ENSMUST00000030840.3
|
Angptl7
|
angiopoietin-like 7 |
| chr19_+_5740885 | 0.53 |
ENSMUST00000081496.5
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr15_-_98607611 | 0.51 |
ENSMUST00000096224.4
|
Adcy6
|
adenylate cyclase 6 |
| chr16_+_17898617 | 0.51 |
ENSMUST00000055374.6
|
Tssk2
|
testis-specific serine kinase 2 |
| chr15_-_103366763 | 0.49 |
ENSMUST00000023128.6
|
Itga5
|
integrin alpha 5 (fibronectin receptor alpha) |
| chr6_-_52240841 | 0.49 |
ENSMUST00000121043.1
|
Hoxa10
|
homeobox A10 |
| chr2_-_93046053 | 0.49 |
ENSMUST00000111272.1
ENSMUST00000178666.1 ENSMUST00000147339.1 |
Prdm11
|
PR domain containing 11 |
| chr1_-_153186447 | 0.47 |
ENSMUST00000027753.6
|
Lamc2
|
laminin, gamma 2 |
| chr12_-_103338314 | 0.47 |
ENSMUST00000149431.1
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr17_+_72918298 | 0.46 |
ENSMUST00000024857.6
|
Lbh
|
limb-bud and heart |
| chr9_+_57940104 | 0.46 |
ENSMUST00000043059.7
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr11_-_116110211 | 0.43 |
ENSMUST00000106441.1
ENSMUST00000021120.5 |
Trim47
|
tripartite motif-containing 47 |
| chr9_+_44072196 | 0.42 |
ENSMUST00000176671.1
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr9_+_106429537 | 0.42 |
ENSMUST00000059802.6
|
Rpl29
|
ribosomal protein L29 |
| chr6_-_112388013 | 0.41 |
ENSMUST00000060847.5
|
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr1_+_153652943 | 0.41 |
ENSMUST00000041776.5
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr1_+_171682004 | 0.41 |
ENSMUST00000015499.7
ENSMUST00000068584.5 |
Cd48
|
CD48 antigen |
| chr11_-_69413675 | 0.39 |
ENSMUST00000094077.4
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chrX_-_104201126 | 0.39 |
ENSMUST00000056502.6
ENSMUST00000118314.1 |
C77370
|
expressed sequence C77370 |
| chrX_+_41401476 | 0.39 |
ENSMUST00000165288.1
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr11_+_115440540 | 0.38 |
ENSMUST00000093914.4
|
4933422H20Rik
|
RIKEN cDNA 4933422H20 gene |
| chr11_+_4160348 | 0.38 |
ENSMUST00000002198.3
|
Sf3a1
|
splicing factor 3a, subunit 1 |
| chr17_-_70851710 | 0.38 |
ENSMUST00000166395.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr8_-_47352348 | 0.36 |
ENSMUST00000110367.2
|
Stox2
|
storkhead box 2 |
| chr2_+_152736244 | 0.36 |
ENSMUST00000038368.8
ENSMUST00000109824.1 |
Id1
|
inhibitor of DNA binding 1 |
| chr10_+_33905015 | 0.36 |
ENSMUST00000169670.1
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr3_-_146839365 | 0.35 |
ENSMUST00000084614.3
|
Gm10288
|
predicted gene 10288 |
| chr7_+_45413657 | 0.35 |
ENSMUST00000058879.6
|
Ntf5
|
neurotrophin 5 |
| chr17_-_35162969 | 0.35 |
ENSMUST00000174805.1
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr4_-_129742275 | 0.35 |
ENSMUST00000066257.5
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr7_+_82867327 | 0.35 |
ENSMUST00000082237.5
|
Mex3b
|
mex3 homolog B (C. elegans) |
| chr9_+_13621646 | 0.35 |
ENSMUST00000034401.8
|
Maml2
|
mastermind like 2 (Drosophila) |
| chr5_+_30913398 | 0.35 |
ENSMUST00000031055.5
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr12_-_36156781 | 0.34 |
ENSMUST00000020856.4
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr18_+_60526194 | 0.34 |
ENSMUST00000025505.5
|
Dctn4
|
dynactin 4 |
| chr3_+_102734496 | 0.33 |
ENSMUST00000029451.5
|
Tspan2
|
tetraspanin 2 |
| chr13_-_98637354 | 0.32 |
ENSMUST00000050389.4
|
Tmem174
|
transmembrane protein 174 |
| chr14_-_25903100 | 0.31 |
ENSMUST00000052286.8
|
Plac9a
|
placenta specific 9a |
| chr3_+_116968267 | 0.30 |
ENSMUST00000117592.1
|
4930455H04Rik
|
RIKEN cDNA 4930455H04 gene |
| chr7_+_4690760 | 0.30 |
ENSMUST00000048248.7
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr2_+_3114220 | 0.29 |
ENSMUST00000072955.5
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr14_-_54554359 | 0.29 |
ENSMUST00000022784.8
|
Haus4
|
HAUS augmin-like complex, subunit 4 |
| chr7_+_101421691 | 0.27 |
ENSMUST00000163751.2
ENSMUST00000084894.7 ENSMUST00000166652.1 |
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated |
| chr8_+_54954728 | 0.27 |
ENSMUST00000033915.7
|
Gpm6a
|
glycoprotein m6a |
| chr14_-_55713088 | 0.26 |
ENSMUST00000002389.7
|
Tgm1
|
transglutaminase 1, K polypeptide |
| chr11_-_102319093 | 0.26 |
ENSMUST00000174302.1
ENSMUST00000178839.1 ENSMUST00000006754.7 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr6_-_138422898 | 0.24 |
ENSMUST00000161450.1
ENSMUST00000163024.1 ENSMUST00000162185.1 |
Lmo3
|
LIM domain only 3 |
| chr2_+_128967383 | 0.24 |
ENSMUST00000110320.2
ENSMUST00000110319.2 |
Zc3h6
|
zinc finger CCCH type containing 6 |
| chr7_+_101361250 | 0.23 |
ENSMUST00000137384.1
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr15_-_38300693 | 0.22 |
ENSMUST00000074043.5
|
Klf10
|
Kruppel-like factor 10 |
| chr15_+_58510037 | 0.20 |
ENSMUST00000161028.1
|
Fer1l6
|
fer-1-like 6 (C. elegans) |
| chr5_+_75574916 | 0.20 |
ENSMUST00000144270.1
ENSMUST00000005815.6 |
Kit
|
kit oncogene |
| chr11_-_103208542 | 0.19 |
ENSMUST00000021323.4
ENSMUST00000107026.2 |
1700023F06Rik
|
RIKEN cDNA 1700023F06 gene |
| chr5_+_64803513 | 0.19 |
ENSMUST00000165536.1
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr13_-_102958084 | 0.19 |
ENSMUST00000099202.3
ENSMUST00000172264.1 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_5295397 | 0.19 |
ENSMUST00000025774.9
|
Sf3b2
|
splicing factor 3b, subunit 2 |
| chr14_+_64950037 | 0.18 |
ENSMUST00000043914.5
|
Ints9
|
integrator complex subunit 9 |
| chr19_-_10738971 | 0.18 |
ENSMUST00000025571.7
|
Cd5
|
CD5 antigen |
| chr7_+_127485221 | 0.18 |
ENSMUST00000048896.6
|
Fbrs
|
fibrosin |
| chr5_+_7179299 | 0.17 |
ENSMUST00000179460.1
|
Tubb4b-ps1
|
tubulin, beta 4B class IVB, pseudogene 1 |
| chr3_+_96221111 | 0.16 |
ENSMUST00000090781.6
|
Hist2h2be
|
histone cluster 2, H2be |
| chr11_-_69122589 | 0.15 |
ENSMUST00000180487.1
|
9130213A22Rik
|
RIKEN cDNA 9130213A22 gene |
| chr4_+_128688726 | 0.15 |
ENSMUST00000106080.1
|
Phc2
|
polyhomeotic-like 2 (Drosophila) |
| chr6_+_38433913 | 0.15 |
ENSMUST00000160583.1
|
Ubn2
|
ubinuclein 2 |
| chr5_+_19907774 | 0.15 |
ENSMUST00000115267.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_-_87101590 | 0.14 |
ENSMUST00000113270.2
|
Alpi
|
alkaline phosphatase, intestinal |
| chr14_+_52835840 | 0.14 |
ENSMUST00000103586.2
|
Trav7d-5
|
T cell receptor alpha variable 7D-5 |
| chr1_-_143702832 | 0.13 |
ENSMUST00000018337.7
|
Cdc73
|
cell division cycle 73, Paf1/RNA polymerase II complex component |
| chrX_-_150588071 | 0.13 |
ENSMUST00000140207.1
ENSMUST00000112719.1 ENSMUST00000112727.3 ENSMUST00000112721.3 ENSMUST00000026303.9 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr11_-_95699143 | 0.13 |
ENSMUST00000062249.2
|
Gm9796
|
predicted gene 9796 |
| chr4_-_134245579 | 0.12 |
ENSMUST00000030644.7
|
Zfp593
|
zinc finger protein 593 |
| chr9_-_36767595 | 0.12 |
ENSMUST00000120381.2
|
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr7_-_30362772 | 0.11 |
ENSMUST00000046351.5
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr6_-_72520538 | 0.11 |
ENSMUST00000089687.4
ENSMUST00000163451.1 ENSMUST00000162561.1 ENSMUST00000159877.1 |
Sh2d6
|
SH2 domain containing 6 |
| chr8_+_95825353 | 0.10 |
ENSMUST00000074053.4
|
Gm10094
|
predicted gene 10094 |
| chr3_+_36552600 | 0.10 |
ENSMUST00000029269.5
ENSMUST00000136890.1 |
Exosc9
|
exosome component 9 |
| chr14_-_20496780 | 0.10 |
ENSMUST00000022353.3
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr14_+_57826210 | 0.10 |
ENSMUST00000022538.3
|
Mrp63
|
mitochondrial ribosomal protein 63 |
| chr14_-_54577578 | 0.10 |
ENSMUST00000054487.8
|
Ajuba
|
ajuba LIM protein |
| chr13_+_111255010 | 0.09 |
ENSMUST00000054716.3
|
Actbl2
|
actin, beta-like 2 |
| chr1_+_153891646 | 0.09 |
ENSMUST00000050660.4
|
Teddm1
|
transmembrane epididymal protein 1 |
| chr1_-_180697034 | 0.08 |
ENSMUST00000027778.7
|
Mixl1
|
Mix1 homeobox-like 1 (Xenopus laevis) |
| chr1_-_64121456 | 0.08 |
ENSMUST00000142009.1
ENSMUST00000114086.1 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_72583245 | 0.08 |
ENSMUST00000145868.1
ENSMUST00000133123.1 ENSMUST00000047615.8 |
Smarcal1
|
SWI/SNF related matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr1_-_54194048 | 0.08 |
ENSMUST00000120904.1
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr15_+_89532816 | 0.08 |
ENSMUST00000167173.1
|
Shank3
|
SH3/ankyrin domain gene 3 |
| chr9_+_96895617 | 0.07 |
ENSMUST00000071781.6
|
Gm10123
|
predicted pseudogene 10123 |
| chrX_-_72274747 | 0.07 |
ENSMUST00000064780.3
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr5_-_24445254 | 0.07 |
ENSMUST00000030800.6
|
Fastk
|
Fas-activated serine/threonine kinase |
| chr19_+_46328179 | 0.06 |
ENSMUST00000026256.2
ENSMUST00000177667.1 |
Fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chr7_+_110221697 | 0.06 |
ENSMUST00000033325.7
|
Swap70
|
SWA-70 protein |
| chr10_-_127534540 | 0.06 |
ENSMUST00000095266.2
|
Nxph4
|
neurexophilin 4 |
| chr7_-_4778141 | 0.05 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr10_+_63457505 | 0.05 |
ENSMUST00000105440.1
|
Ctnna3
|
catenin (cadherin associated protein), alpha 3 |
| chr5_-_24445166 | 0.05 |
ENSMUST00000115043.1
ENSMUST00000115041.1 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr7_+_45785331 | 0.05 |
ENSMUST00000120005.1
ENSMUST00000123585.1 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr14_+_50944499 | 0.04 |
ENSMUST00000178092.1
|
Pnp
|
purine-nucleoside phosphorylase |
| chr19_+_4231899 | 0.04 |
ENSMUST00000025773.3
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chrX_+_99136119 | 0.03 |
ENSMUST00000052839.6
|
Efnb1
|
ephrin B1 |
| chr6_+_108660772 | 0.03 |
ENSMUST00000163617.1
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr1_-_75232093 | 0.03 |
ENSMUST00000180101.1
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr14_+_55510445 | 0.02 |
ENSMUST00000165262.1
ENSMUST00000074225.4 |
Cpne6
|
copine VI |
| chr2_-_128967725 | 0.02 |
ENSMUST00000099385.2
|
Gm10762
|
predicted gene 10762 |
| chr11_+_53433299 | 0.02 |
ENSMUST00000018382.6
|
Gdf9
|
growth differentiation factor 9 |
| chr7_+_128203598 | 0.02 |
ENSMUST00000177383.1
|
Itgad
|
integrin, alpha D |
| chr10_-_92164666 | 0.02 |
ENSMUST00000183123.1
ENSMUST00000182033.1 |
Rmst
|
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
| chr4_+_99272671 | 0.01 |
ENSMUST00000094956.1
|
Gm10305
|
predicted gene 10305 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.8 | 4.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.6 | 1.9 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.5 | 1.4 | GO:1905204 | negative regulation of connective tissue replacement(GO:1905204) positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.5 | 1.4 | GO:0070315 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.5 | 2.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 2.5 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 1.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 0.7 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.3 | 0.9 | GO:0045763 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.3 | 0.8 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.2 | 0.7 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.2 | 2.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 4.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 1.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 0.5 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 1.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.7 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 1.7 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 1.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.7 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.4 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.4 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 1.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.7 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 1.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 1.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.5 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.4 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 1.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 1.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.2 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 1.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.9 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.5 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 1.0 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 1.6 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 1.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.6 | 1.7 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 4.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 4.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.5 | GO:0005610 | laminin-2 complex(GO:0005607) laminin-5 complex(GO:0005610) |
| 0.1 | 1.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.8 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.3 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.6 | 1.7 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.6 | 1.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.5 | 2.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 2.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 4.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 0.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 1.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.7 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 7.8 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |