Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
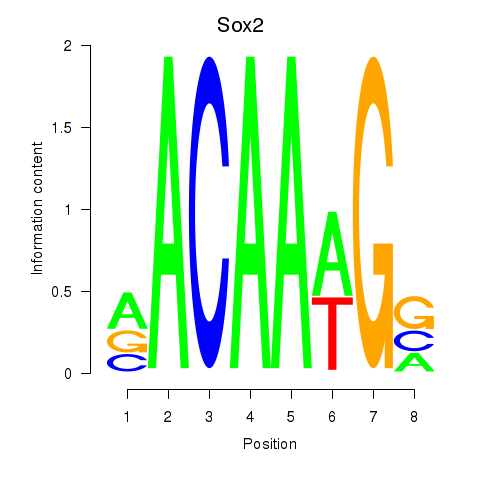
Results for Sox2
Z-value: 2.61
Transcription factors associated with Sox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox2
|
ENSMUSG00000074637.4 | SRY (sex determining region Y)-box 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox2 | mm10_v2_chr3_+_34649987_34650005 | 0.40 | 1.7e-02 | Click! |
Activity profile of Sox2 motif
Sorted Z-values of Sox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_134915512 | 25.47 |
ENSMUST00000008987.4
|
Cldn13
|
claudin 13 |
| chr10_+_43579161 | 23.12 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr7_+_45216671 | 21.50 |
ENSMUST00000134420.1
|
Tead2
|
TEA domain family member 2 |
| chr4_+_115057683 | 21.44 |
ENSMUST00000161601.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr7_-_142661305 | 18.19 |
ENSMUST00000105936.1
|
Igf2
|
insulin-like growth factor 2 |
| chr11_+_116531097 | 15.02 |
ENSMUST00000138840.1
|
Sphk1
|
sphingosine kinase 1 |
| chr16_-_36367623 | 13.41 |
ENSMUST00000096089.2
|
BC100530
|
cDNA sequence BC100530 |
| chr16_-_17125106 | 12.91 |
ENSMUST00000093336.6
|
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr3_-_98859774 | 12.51 |
ENSMUST00000107016.3
ENSMUST00000149768.1 |
Hsd3b1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr3_-_98859753 | 11.87 |
ENSMUST00000029465.6
|
Hsd3b1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr6_+_86628174 | 11.66 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr5_+_44100442 | 11.52 |
ENSMUST00000072800.4
|
Gm16401
|
predicted gene 16401 |
| chr9_-_103480328 | 11.48 |
ENSMUST00000124310.2
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr18_+_82554463 | 11.14 |
ENSMUST00000062446.7
ENSMUST00000102812.4 ENSMUST00000075372.5 ENSMUST00000080658.4 ENSMUST00000152071.1 ENSMUST00000114674.3 ENSMUST00000142850.1 ENSMUST00000133193.1 ENSMUST00000123251.1 ENSMUST00000153478.1 ENSMUST00000132369.1 |
Mbp
|
myelin basic protein |
| chr6_-_83527452 | 10.86 |
ENSMUST00000141904.1
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr1_-_132390301 | 10.69 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr4_+_115057410 | 10.53 |
ENSMUST00000136946.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr3_-_84480419 | 10.29 |
ENSMUST00000107689.1
|
Fhdc1
|
FH2 domain containing 1 |
| chr7_-_103853199 | 10.27 |
ENSMUST00000033229.3
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr2_+_84734050 | 9.97 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr15_-_103251465 | 9.29 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr4_+_141010644 | 9.20 |
ENSMUST00000071977.8
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr17_-_79355082 | 9.17 |
ENSMUST00000068958.7
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr11_-_96005872 | 9.15 |
ENSMUST00000013559.2
|
Igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr1_+_136131382 | 9.14 |
ENSMUST00000075164.4
|
Kif21b
|
kinesin family member 21B |
| chr12_+_24831583 | 9.12 |
ENSMUST00000110942.3
ENSMUST00000078902.6 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr10_-_21160925 | 9.07 |
ENSMUST00000020158.6
|
Myb
|
myeloblastosis oncogene |
| chr4_+_136172367 | 9.06 |
ENSMUST00000061721.5
|
E2f2
|
E2F transcription factor 2 |
| chr13_+_117220584 | 9.05 |
ENSMUST00000022242.7
|
Emb
|
embigin |
| chr4_-_148130678 | 9.01 |
ENSMUST00000030862.4
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr7_-_120982260 | 8.96 |
ENSMUST00000033169.8
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr9_+_58014990 | 8.90 |
ENSMUST00000034874.7
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr2_+_156840966 | 8.82 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr3_-_20275659 | 8.64 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr12_-_36042476 | 8.53 |
ENSMUST00000020896.8
|
Tspan13
|
tetraspanin 13 |
| chr4_+_62965560 | 8.37 |
ENSMUST00000030043.6
ENSMUST00000107415.1 ENSMUST00000064814.5 |
Zfp618
|
zinc finger protein 618 |
| chr11_+_58948890 | 8.36 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr14_+_80000292 | 8.35 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr17_+_48232755 | 8.28 |
ENSMUST00000113251.3
ENSMUST00000048782.6 |
Trem1
|
triggering receptor expressed on myeloid cells 1 |
| chr3_-_54915867 | 8.15 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr11_+_78301529 | 8.12 |
ENSMUST00000045026.3
|
Spag5
|
sperm associated antigen 5 |
| chr2_-_168767136 | 8.06 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr7_+_45215753 | 8.04 |
ENSMUST00000033060.6
ENSMUST00000155313.1 ENSMUST00000107801.1 |
Tead2
|
TEA domain family member 2 |
| chr11_+_11686213 | 7.82 |
ENSMUST00000076700.4
ENSMUST00000048122.6 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr15_-_103255433 | 7.82 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chrX_+_93654863 | 7.71 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr2_+_155611175 | 7.65 |
ENSMUST00000092995.5
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr15_-_36879816 | 7.51 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr2_-_170406501 | 7.48 |
ENSMUST00000154650.1
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr2_+_131491958 | 7.45 |
ENSMUST00000110181.1
ENSMUST00000110180.1 |
Smox
|
spermine oxidase |
| chr4_-_130574150 | 7.41 |
ENSMUST00000105993.3
|
Nkain1
|
Na+/K+ transporting ATPase interacting 1 |
| chr5_-_44099220 | 7.38 |
ENSMUST00000165909.1
|
Prom1
|
prominin 1 |
| chr2_+_131491764 | 7.36 |
ENSMUST00000028806.5
ENSMUST00000110179.2 ENSMUST00000110189.2 ENSMUST00000110182.2 ENSMUST00000110183.2 ENSMUST00000110186.2 ENSMUST00000110188.1 |
Smox
|
spermine oxidase |
| chr9_+_111004811 | 7.27 |
ENSMUST00000080872.4
|
Gm10030
|
predicted gene 10030 |
| chr2_+_4559742 | 7.23 |
ENSMUST00000176828.1
|
Frmd4a
|
FERM domain containing 4A |
| chr12_+_24708984 | 7.15 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr15_-_96642883 | 7.14 |
ENSMUST00000088452.4
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr17_+_12119274 | 7.10 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr7_+_100493337 | 7.04 |
ENSMUST00000126534.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_+_62077018 | 7.00 |
ENSMUST00000092415.5
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr13_-_55488038 | 6.99 |
ENSMUST00000109921.2
ENSMUST00000109923.2 ENSMUST00000021950.8 |
Dbn1
|
drebrin 1 |
| chr7_-_116038734 | 6.93 |
ENSMUST00000166877.1
|
Sox6
|
SRY-box containing gene 6 |
| chr8_+_123332676 | 6.85 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr11_+_11685909 | 6.74 |
ENSMUST00000065433.5
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr2_+_120476911 | 6.66 |
ENSMUST00000110716.1
ENSMUST00000028748.6 ENSMUST00000090028.5 ENSMUST00000110719.2 |
Capn3
|
calpain 3 |
| chr6_-_87590701 | 6.64 |
ENSMUST00000050887.7
|
Prokr1
|
prokineticin receptor 1 |
| chr8_+_123411424 | 6.60 |
ENSMUST00000071134.3
|
Tubb3
|
tubulin, beta 3 class III |
| chr17_-_35000848 | 6.60 |
ENSMUST00000166828.3
|
D17H6S56E-5
|
DNA segment, Chr 17, human D6S56E 5 |
| chrX_+_93675088 | 6.55 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chrX_+_134295225 | 6.52 |
ENSMUST00000037687.7
|
Tmem35
|
transmembrane protein 35 |
| chr5_-_138171248 | 6.48 |
ENSMUST00000153867.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr10_-_30655859 | 6.47 |
ENSMUST00000092610.4
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr10_-_122047293 | 6.44 |
ENSMUST00000020322.5
ENSMUST00000081688.6 |
Srgap1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr7_+_100493795 | 6.43 |
ENSMUST00000129324.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr1_-_182019927 | 6.38 |
ENSMUST00000078719.6
ENSMUST00000111030.3 ENSMUST00000177811.1 ENSMUST00000111024.3 ENSMUST00000111025.2 |
Enah
|
enabled homolog (Drosophila) |
| chr5_-_108749448 | 6.37 |
ENSMUST00000068946.7
|
Rnf212
|
ring finger protein 212 |
| chrX_+_21484532 | 6.32 |
ENSMUST00000089188.2
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr7_-_100514800 | 6.25 |
ENSMUST00000054923.7
|
Dnajb13
|
DnaJ (Hsp40) related, subfamily B, member 13 |
| chr7_-_115824699 | 6.21 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr2_-_168767029 | 6.15 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr16_+_18127607 | 6.10 |
ENSMUST00000059589.5
|
Rtn4r
|
reticulon 4 receptor |
| chr6_-_83527773 | 6.08 |
ENSMUST00000152029.1
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr18_+_34624621 | 6.08 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr5_-_138170992 | 6.02 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr7_+_141061274 | 6.00 |
ENSMUST00000048002.5
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr10_-_37138863 | 6.00 |
ENSMUST00000092584.5
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr11_-_69560186 | 6.00 |
ENSMUST00000004036.5
|
Efnb3
|
ephrin B3 |
| chr5_-_131538687 | 5.98 |
ENSMUST00000161374.1
|
Auts2
|
autism susceptibility candidate 2 |
| chr13_+_52583437 | 5.96 |
ENSMUST00000118756.1
|
Syk
|
spleen tyrosine kinase |
| chr12_-_54986363 | 5.96 |
ENSMUST00000173433.1
ENSMUST00000173803.1 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr11_+_67277124 | 5.95 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr1_+_74409376 | 5.93 |
ENSMUST00000027366.6
|
Vil1
|
villin 1 |
| chr1_+_153749496 | 5.93 |
ENSMUST00000182722.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr4_-_83021102 | 5.92 |
ENSMUST00000071708.5
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr3_+_14886426 | 5.92 |
ENSMUST00000029078.7
|
Car2
|
carbonic anhydrase 2 |
| chr17_-_56476462 | 5.91 |
ENSMUST00000067538.5
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr12_-_54986328 | 5.89 |
ENSMUST00000038926.6
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr11_+_32276400 | 5.87 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr8_-_85365341 | 5.84 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr11_+_3649759 | 5.83 |
ENSMUST00000140242.1
|
Morc2a
|
microrchidia 2A |
| chr2_-_131160006 | 5.81 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr11_+_95010277 | 5.77 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr3_+_88615367 | 5.77 |
ENSMUST00000176539.1
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr8_-_85365317 | 5.73 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr14_+_60378242 | 5.72 |
ENSMUST00000022561.6
|
Amer2
|
APC membrane recruitment 2 |
| chr11_-_46312220 | 5.68 |
ENSMUST00000129474.1
ENSMUST00000093166.4 ENSMUST00000165599.2 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr16_+_11066292 | 5.62 |
ENSMUST00000089011.4
|
Snn
|
stannin |
| chr7_-_143460989 | 5.62 |
ENSMUST00000167912.1
ENSMUST00000037287.6 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr5_-_138171216 | 5.62 |
ENSMUST00000147920.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr2_-_164389095 | 5.59 |
ENSMUST00000167427.1
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr11_-_74590065 | 5.56 |
ENSMUST00000145524.1
ENSMUST00000047488.7 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr4_+_107830958 | 5.56 |
ENSMUST00000106731.2
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr18_+_34625009 | 5.55 |
ENSMUST00000166044.1
|
Kif20a
|
kinesin family member 20A |
| chr2_+_157560078 | 5.52 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr3_+_153973436 | 5.48 |
ENSMUST00000089948.5
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr8_-_69184177 | 5.47 |
ENSMUST00000185176.1
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr11_-_87108656 | 5.47 |
ENSMUST00000051395.8
|
Prr11
|
proline rich 11 |
| chr2_-_101883010 | 5.43 |
ENSMUST00000154525.1
|
Prr5l
|
proline rich 5 like |
| chr7_-_84409959 | 5.43 |
ENSMUST00000085077.3
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr19_-_5894100 | 5.41 |
ENSMUST00000055911.4
|
Tigd3
|
tigger transposable element derived 3 |
| chr18_-_23981555 | 5.39 |
ENSMUST00000115829.1
|
Zscan30
|
zinc finger and SCAN domain containing 30 |
| chr15_-_84557776 | 5.39 |
ENSMUST00000069476.4
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like |
| chr7_+_110777653 | 5.37 |
ENSMUST00000148292.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chrX_-_164258186 | 5.33 |
ENSMUST00000112265.2
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr16_-_20426375 | 5.32 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr10_-_80421847 | 5.31 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr7_-_142095266 | 5.29 |
ENSMUST00000039926.3
|
Dusp8
|
dual specificity phosphatase 8 |
| chr19_-_46039621 | 5.28 |
ENSMUST00000056931.7
|
Ldb1
|
LIM domain binding 1 |
| chr2_+_31950257 | 5.25 |
ENSMUST00000001920.7
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr4_-_154928545 | 5.25 |
ENSMUST00000152687.1
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr8_+_94977101 | 5.22 |
ENSMUST00000179619.1
|
Gpr56
|
G protein-coupled receptor 56 |
| chr2_-_84822546 | 5.21 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chrX_+_48519245 | 5.17 |
ENSMUST00000033430.2
|
Rab33a
|
RAB33A, member of RAS oncogene family |
| chrX_-_59568068 | 5.15 |
ENSMUST00000119833.1
ENSMUST00000131319.1 |
Fgf13
|
fibroblast growth factor 13 |
| chr11_+_95009852 | 5.14 |
ENSMUST00000055947.3
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr1_-_93342734 | 5.13 |
ENSMUST00000027493.3
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr11_+_69846610 | 5.10 |
ENSMUST00000152566.1
ENSMUST00000108633.2 |
Plscr3
|
phospholipid scramblase 3 |
| chr4_-_132049058 | 5.09 |
ENSMUST00000105981.2
ENSMUST00000084253.3 ENSMUST00000141291.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chrX_-_143827391 | 5.08 |
ENSMUST00000087316.5
|
Capn6
|
calpain 6 |
| chr10_+_41476314 | 4.98 |
ENSMUST00000119962.1
ENSMUST00000019967.9 ENSMUST00000099934.4 |
Mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr17_-_48409729 | 4.98 |
ENSMUST00000160319.1
ENSMUST00000159535.1 ENSMUST00000078800.6 ENSMUST00000046719.7 ENSMUST00000162460.1 |
Nfya
|
nuclear transcription factor-Y alpha |
| chr11_-_74590186 | 4.96 |
ENSMUST00000102521.1
|
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr6_-_88875035 | 4.95 |
ENSMUST00000145944.1
|
Podxl2
|
podocalyxin-like 2 |
| chr7_+_45335256 | 4.93 |
ENSMUST00000085351.4
|
Hrc
|
histidine rich calcium binding protein |
| chr12_+_108605757 | 4.92 |
ENSMUST00000109854.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr7_+_24897381 | 4.91 |
ENSMUST00000003469.7
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr8_+_86745679 | 4.90 |
ENSMUST00000098532.2
|
Gm10638
|
predicted gene 10638 |
| chr3_-_89338005 | 4.89 |
ENSMUST00000029674.7
|
Efna4
|
ephrin A4 |
| chr6_-_67037399 | 4.88 |
ENSMUST00000043098.6
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr16_+_17233560 | 4.88 |
ENSMUST00000090190.5
ENSMUST00000115698.2 |
Hic2
|
hypermethylated in cancer 2 |
| chr1_+_87404916 | 4.87 |
ENSMUST00000173152.1
ENSMUST00000173663.1 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr2_-_114052804 | 4.84 |
ENSMUST00000090269.6
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr12_+_24708241 | 4.83 |
ENSMUST00000020980.5
|
Rrm2
|
ribonucleotide reductase M2 |
| chr6_+_127887582 | 4.83 |
ENSMUST00000032501.4
|
Tspan11
|
tetraspanin 11 |
| chr6_-_87335758 | 4.81 |
ENSMUST00000042025.9
|
Antxr1
|
anthrax toxin receptor 1 |
| chr5_+_105415738 | 4.80 |
ENSMUST00000112707.1
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr12_-_101028983 | 4.80 |
ENSMUST00000068411.3
ENSMUST00000085096.3 |
Ccdc88c
|
coiled-coil domain containing 88C |
| chr1_+_153749414 | 4.75 |
ENSMUST00000086209.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr6_-_148944750 | 4.74 |
ENSMUST00000111562.1
ENSMUST00000081956.5 |
Fam60a
|
family with sequence similarity 60, member A |
| chr4_+_43957678 | 4.74 |
ENSMUST00000107855.1
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr4_-_134012381 | 4.73 |
ENSMUST00000176113.1
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr7_-_113347273 | 4.73 |
ENSMUST00000117577.1
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr11_+_69846665 | 4.72 |
ENSMUST00000019605.2
|
Plscr3
|
phospholipid scramblase 3 |
| chr1_+_139454747 | 4.72 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr5_+_26904682 | 4.68 |
ENSMUST00000120555.1
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr4_-_63403330 | 4.63 |
ENSMUST00000035724.4
|
Akna
|
AT-hook transcription factor |
| chr5_+_66676098 | 4.59 |
ENSMUST00000031131.9
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr12_-_16800674 | 4.57 |
ENSMUST00000162112.1
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr11_-_102819114 | 4.56 |
ENSMUST00000068933.5
|
Gjc1
|
gap junction protein, gamma 1 |
| chr6_+_128375456 | 4.54 |
ENSMUST00000100926.2
|
4933413G19Rik
|
RIKEN cDNA 4933413G19 gene |
| chr9_+_21029373 | 4.51 |
ENSMUST00000001040.5
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr5_+_37245792 | 4.46 |
ENSMUST00000031004.7
|
Crmp1
|
collapsin response mediator protein 1 |
| chr11_+_16951371 | 4.46 |
ENSMUST00000109635.1
ENSMUST00000061327.1 |
Fbxo48
|
F-box protein 48 |
| chr16_-_20426322 | 4.43 |
ENSMUST00000115547.2
ENSMUST00000096199.4 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr16_-_20425881 | 4.41 |
ENSMUST00000077867.3
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_-_25005895 | 4.36 |
ENSMUST00000102858.3
ENSMUST00000080882.6 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr19_+_44293676 | 4.34 |
ENSMUST00000026221.5
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr18_-_43737186 | 4.28 |
ENSMUST00000025381.2
|
Spink3
|
serine peptidase inhibitor, Kazal type 3 |
| chr14_+_31019183 | 4.27 |
ENSMUST00000052239.5
|
Pbrm1
|
polybromo 1 |
| chr4_-_58499398 | 4.27 |
ENSMUST00000107570.1
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr6_+_40964760 | 4.24 |
ENSMUST00000076638.5
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr12_-_115790884 | 4.23 |
ENSMUST00000081809.5
|
Ighv1-73
|
immunoglobulin heavy variable 1-73 |
| chr11_+_96323253 | 4.22 |
ENSMUST00000093944.3
|
Hoxb3
|
homeobox B3 |
| chr5_+_142702091 | 4.22 |
ENSMUST00000058418.7
|
Slc29a4
|
solute carrier family 29 (nucleoside transporters), member 4 |
| chr8_+_109705549 | 4.20 |
ENSMUST00000034163.8
|
Zfp821
|
zinc finger protein 821 |
| chr18_+_82475133 | 4.16 |
ENSMUST00000091789.4
ENSMUST00000114676.1 ENSMUST00000047865.7 |
Mbp
|
myelin basic protein |
| chr15_+_80173642 | 4.15 |
ENSMUST00000044970.6
|
Mgat3
|
mannoside acetylglucosaminyltransferase 3 |
| chr10_+_128908907 | 4.15 |
ENSMUST00000105229.1
|
Cd63
|
CD63 antigen |
| chr11_+_87760533 | 4.14 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr19_-_47919269 | 4.13 |
ENSMUST00000095998.5
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr10_-_79874233 | 4.12 |
ENSMUST00000166023.1
ENSMUST00000167707.1 ENSMUST00000165601.1 |
BC005764
|
cDNA sequence BC005764 |
| chr10_+_17796256 | 4.11 |
ENSMUST00000037964.6
|
Txlnb
|
taxilin beta |
| chr16_+_19760902 | 4.11 |
ENSMUST00000119468.1
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr16_-_18629864 | 4.11 |
ENSMUST00000096987.5
|
Sept5
|
septin 5 |
| chr17_-_25433775 | 4.10 |
ENSMUST00000159610.1
ENSMUST00000159048.1 ENSMUST00000078496.5 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr4_-_128806045 | 4.10 |
ENSMUST00000106072.2
ENSMUST00000170934.1 |
Zfp362
|
zinc finger protein 362 |
| chr4_+_48585193 | 4.09 |
ENSMUST00000107703.1
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr4_+_107802277 | 4.08 |
ENSMUST00000106733.2
ENSMUST00000030356.3 ENSMUST00000106732.2 ENSMUST00000126573.1 |
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr8_+_36094828 | 4.08 |
ENSMUST00000110492.1
|
D8Ertd82e
|
DNA segment, Chr 8, ERATO Doi 82, expressed |
| chr5_+_134676490 | 4.04 |
ENSMUST00000100641.2
|
Gm10369
|
predicted gene 10369 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 7.7 | 23.1 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 5.1 | 15.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 4.9 | 14.6 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 2.5 | 7.5 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 2.5 | 15.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 2.5 | 7.4 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 2.4 | 11.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 2.3 | 6.8 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 2.3 | 18.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.2 | 9.0 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 2.2 | 6.7 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 2.2 | 6.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 2.1 | 6.4 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 2.1 | 14.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 2.1 | 6.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 2.0 | 16.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.0 | 6.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.0 | 5.9 | GO:0035672 | positive regulation of cellular pH reduction(GO:0032849) oligopeptide transmembrane transport(GO:0035672) |
| 2.0 | 7.8 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 1.8 | 18.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.8 | 9.0 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 1.8 | 10.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 1.8 | 7.0 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 1.6 | 8.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 1.6 | 9.6 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 1.6 | 6.3 | GO:1904009 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 1.5 | 6.2 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.5 | 6.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 1.5 | 4.6 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 1.5 | 4.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 1.5 | 6.0 | GO:0042223 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 1.5 | 5.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.4 | 17.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 1.4 | 12.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 1.3 | 16.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 1.3 | 9.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.3 | 3.9 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.3 | 3.9 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 1.3 | 9.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.3 | 3.9 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 1.3 | 2.6 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.3 | 3.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.3 | 6.4 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 1.3 | 10.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 1.2 | 3.7 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 1.2 | 4.9 | GO:0033374 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 1.2 | 7.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 1.2 | 8.3 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 1.2 | 5.9 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 1.2 | 4.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 1.2 | 9.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.1 | 30.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 1.1 | 4.5 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.1 | 4.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.1 | 18.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 1.1 | 3.2 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 1.1 | 4.3 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.1 | 4.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 1.0 | 8.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.0 | 3.8 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 1.0 | 4.8 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.0 | 5.7 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.9 | 2.8 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.9 | 11.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.9 | 8.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.9 | 2.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.9 | 4.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.9 | 2.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.9 | 5.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.9 | 7.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.9 | 12.8 | GO:0033275 | actin-myosin filament sliding(GO:0033275) |
| 0.8 | 2.5 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.8 | 15.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.8 | 4.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.8 | 8.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.8 | 11.3 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.8 | 4.8 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.8 | 7.9 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.8 | 7.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.8 | 5.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.8 | 4.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.8 | 3.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.8 | 14.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.7 | 1.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.7 | 3.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.7 | 0.7 | GO:0060529 | squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.7 | 1.4 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.7 | 2.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.7 | 3.6 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.7 | 2.8 | GO:0035127 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.7 | 0.7 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 | 5.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 2.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.7 | 3.5 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.7 | 2.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.7 | 2.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.7 | 3.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.7 | 3.4 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.7 | 6.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.7 | 2.0 | GO:0055130 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.6 | 6.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.6 | 3.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.6 | 5.1 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.6 | 3.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.6 | 14.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.6 | 2.5 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.6 | 10.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.6 | 1.9 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.6 | 4.9 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.6 | 11.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.6 | 5.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.6 | 10.8 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) |
| 0.6 | 1.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.6 | 5.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 1.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.6 | 13.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.6 | 2.3 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.6 | 1.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.6 | 2.8 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.6 | 2.3 | GO:0060032 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) notochord regression(GO:0060032) |
| 0.6 | 3.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.5 | 7.1 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.5 | 4.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 2.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 5.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.5 | 2.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.5 | 3.6 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.5 | 2.1 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.5 | 1.5 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.5 | 2.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.5 | 5.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 2.5 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.5 | 1.5 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.5 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 2.4 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.5 | 4.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.5 | 6.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.5 | 1.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.5 | 8.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 6.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 1.4 | GO:0090260 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance through spinal cord(GO:0021972) regulation of negative chemotaxis(GO:0050923) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.5 | 1.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.4 | 1.8 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.4 | 2.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.4 | 2.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 11.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.4 | 4.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.4 | 2.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 9.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.4 | 2.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.4 | 10.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.4 | 1.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.4 | 6.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 1.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 1.1 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.4 | 3.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.4 | 8.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.4 | 3.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.4 | 8.8 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.4 | 9.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 1.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 11.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.4 | 1.8 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 7.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 2.9 | GO:0040016 | DNA topological change(GO:0006265) embryonic cleavage(GO:0040016) |
| 0.4 | 12.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.4 | 21.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.4 | 6.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.4 | 0.7 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.3 | 1.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.3 | 2.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 1.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 1.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 1.0 | GO:0048319 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) axial mesoderm morphogenesis(GO:0048319) |
| 0.3 | 1.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.3 | 3.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 1.0 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 1.6 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 1.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 0.6 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.3 | 3.5 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 1.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 1.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 3.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 3.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.3 | 6.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.3 | 3.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 0.9 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 | 0.6 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.3 | 2.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 | 3.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.3 | 2.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 0.8 | GO:0002631 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.3 | 2.4 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.3 | 2.6 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 2.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 0.3 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.3 | 1.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 0.8 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.3 | 0.8 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.3 | 2.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.3 | 2.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.2 | 0.7 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 1.2 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.2 | 3.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.0 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.2 | 1.0 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 5.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 10.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 1.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 2.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 0.9 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 0.9 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 3.2 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.2 | 0.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 15.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.2 | 1.3 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 | 2.7 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 4.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.2 | 2.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 1.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 1.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 1.3 | GO:0009624 | response to nematode(GO:0009624) |
| 0.2 | 16.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 5.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 0.6 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 1.8 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.2 | 1.4 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.2 | 3.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 1.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 3.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 1.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 4.9 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.2 | 1.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 7.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 4.7 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.2 | 3.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 1.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 1.9 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.2 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 1.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 8.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 2.7 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.2 | 0.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 1.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 3.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 1.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.2 | 1.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 2.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 2.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.5 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 3.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 3.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 5.0 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 | 1.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 4.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.2 | 0.7 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.2 | 1.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.2 | 0.7 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 1.0 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.2 | 1.5 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.2 | 1.6 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.2 | 1.9 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.2 | 6.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 0.6 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 7.5 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.2 | 1.4 | GO:0014824 | artery smooth muscle contraction(GO:0014824) positive regulation of odontogenesis(GO:0042482) |
| 0.2 | 3.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 5.2 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 1.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 3.0 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 3.2 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.1 | 0.1 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 4.8 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 1.2 | GO:0048619 | hindgut morphogenesis(GO:0007442) embryonic genitalia morphogenesis(GO:0030538) embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 1.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 1.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.4 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 2.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.8 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.4 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.1 | 0.4 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 2.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 1.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 2.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 3.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 4.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.1 | 0.3 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 0.3 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 1.7 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.4 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.1 | 10.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.3 | GO:2000020 | positive regulation of male gonad development(GO:2000020) negative regulation of female gonad development(GO:2000195) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.5 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 0.8 | GO:0071816 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 1.5 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 5.6 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 2.7 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 1.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.1 | 0.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 10.7 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 0.3 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 5.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.1 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.0 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.1 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 2.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 3.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 1.0 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 4.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.0 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 12.8 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 0.1 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 3.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 2.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 5.5 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 1.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 3.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 2.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.3 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 1.3 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 0.4 | GO:1904645 | response to beta-amyloid(GO:1904645) |
| 0.1 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 3.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.3 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 3.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:1902022 | L-lysine transport(GO:1902022) |
| 0.0 | 1.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 2.2 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 4.4 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.2 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.3 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 1.3 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 3.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.3 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.4 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 2.6 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.2 | GO:0046856 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.6 | GO:0098840 | protein transport along microtubule(GO:0098840) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 9.8 | 29.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 3.0 | 11.8 | GO:0008623 | CHRAC(GO:0008623) |
| 2.4 | 12.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 2.0 | 16.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.0 | 6.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 1.9 | 13.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.8 | 10.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.7 | 7.0 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 1.7 | 1.7 | GO:0035101 | FACT complex(GO:0035101) |
| 1.6 | 4.9 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 1.5 | 7.4 | GO:0071914 | prominosome(GO:0071914) |
| 1.3 | 6.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.2 | 2.5 | GO:0044393 | microspike(GO:0044393) |
| 1.2 | 9.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.2 | 26.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.1 | 16.9 | GO:0042555 | MCM complex(GO:0042555) |
| 1.1 | 3.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.1 | 18.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.1 | 11.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 1.0 | 4.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.0 | 8.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.0 | 6.8 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 1.0 | 5.7 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.9 | 9.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.9 | 3.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.9 | 1.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.8 | 11.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 7.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.8 | 2.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 2.0 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.6 | 1.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.6 | 9.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 7.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.6 | 2.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.6 | 1.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.6 | 2.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.6 | 1.8 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.6 | 1.7 | GO:0005593 | FACIT collagen trimer(GO:0005593) collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.6 | 8.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.5 | 3.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.5 | 8.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.5 | 3.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 8.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.5 | 7.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 1.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.5 | 8.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.5 | 2.9 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.5 | 3.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 2.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 4.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 4.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.4 | 5.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.4 | 1.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 2.9 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.4 | 7.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 2.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 2.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.4 | 2.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.4 | 7.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 1.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 12.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.4 | 2.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 3.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 5.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 2.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 3.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 5.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 2.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 8.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 0.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 1.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 9.4 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 5.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 5.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 0.8 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 6.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 4.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 0.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 1.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 0.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 7.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 14.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 2.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 1.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 4.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 35.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 11.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 2.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 2.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 1.3 | GO:0070160 | occluding junction(GO:0070160) |
| 0.2 | 4.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 17.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 2.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 1.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 2.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 1.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.3 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 1.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 5.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 16.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 3.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.6 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 4.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 7.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 13.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 5.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 10.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 10.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 4.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 24.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.9 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 6.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 0.8 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 4.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 3.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 5.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 3.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 9.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 5.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 5.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 3.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 21.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 3.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 7.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 1.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 1.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 3.0 | 24.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 2.9 | 14.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 2.8 | 16.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 2.6 | 10.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 2.4 | 9.6 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 2.4 | 12.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.3 | 9.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 2.2 | 11.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 2.1 | 15.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 2.1 | 6.4 | GO:0004349 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.7 | 33.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 1.7 | 6.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.7 | 11.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.6 | 6.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 1.5 | 9.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 1.5 | 19.7 | GO:0005522 | profilin binding(GO:0005522) |
| 1.5 | 13.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.4 | 7.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 1.4 | 4.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.3 | 10.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.3 | 21.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 1.3 | 10.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.3 | 2.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 1.2 | 7.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.2 | 4.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.1 | 4.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.1 | 4.6 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 1.1 | 3.4 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 1.1 | 6.6 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 1.1 | 4.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.1 | 3.3 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 1.1 | 4.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 1.0 | 5.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.0 | 7.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.0 | 5.7 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.9 | 8.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.9 | 3.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.9 | 3.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.9 | 3.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.8 | 2.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.8 | 3.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.8 | 4.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.8 | 5.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.7 | 5.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.7 | 2.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.7 | 3.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.7 | 2.9 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.7 | 2.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.7 | 7.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.7 | 9.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 2.1 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.7 | 2.8 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.7 | 2.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.7 | 2.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.7 | 2.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.7 | 7.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.6 | 36.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.6 | 1.9 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.6 | 1.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.6 | 5.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.6 | 5.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.6 | 1.7 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.6 | 2.3 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.6 | 2.8 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.6 | 2.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.6 | 5.5 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.6 | 0.6 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.6 | 3.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 3.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.6 | 19.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.6 | 5.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.5 | 15.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.5 | 17.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.5 | 3.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.5 | 6.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 7.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 2.0 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 11.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.5 | 2.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 16.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.5 | 4.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 2.8 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.5 | 1.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 3.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 7.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 1.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.4 | 10.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.4 | 4.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.4 | 2.1 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 16.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.4 | 2.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.4 | 2.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 1.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.4 | 1.6 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.4 | 3.5 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 3.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 2.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.4 | 8.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 1.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.4 | 2.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 1.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 3.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.4 | 3.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.4 | 3.9 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.4 | 7.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 0.7 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 2.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 4.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 7.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 3.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 1.6 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.3 | 3.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 12.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 4.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 1.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 9.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 3.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 2.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 6.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 6.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 2.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 13.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 1.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 2.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 6.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 2.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 2.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 8.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.3 | 6.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 2.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 1.7 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 1.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.2 | 3.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 3.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 1.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 1.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 1.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 7.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 1.6 | GO:0046935 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 7.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 5.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 15.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 2.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 4.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 4.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 1.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 0.7 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.2 | 3.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 7.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 3.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 10.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 1.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 8.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 7.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 8.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 6.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 1.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 10.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.7 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 9.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 2.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 14.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 7.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 6.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.4 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 2.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 4.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 2.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 6.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 3.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 3.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 12.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 1.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 2.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 4.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.3 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 2.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.6 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 8.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.8 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 6.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 19.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 2.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 4.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.2 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 2.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 1.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.8 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 3.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 7.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.9 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 4.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 3.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 7.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 15.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.5 | 30.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.5 | 7.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 8.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 17.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 6.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.4 | 9.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.4 | 21.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 4.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 14.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 23.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 10.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 5.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 3.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 11.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 21.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 7.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 12.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 9.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 7.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 3.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 5.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 3.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 3.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 8.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 4.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 3.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 5.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 5.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 2.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 5.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 0.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 5.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 6.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 9.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 5.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 4.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 24.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 1.2 | 19.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.0 | 7.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.0 | 16.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.9 | 18.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 7.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.7 | 13.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.7 | 11.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.7 | 12.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.6 | 19.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.6 | 24.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.6 | 11.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 6.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 10.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.5 | 12.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 12.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.4 | 16.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 17.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 15.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 8.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 13.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 9.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.3 | 1.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.3 | 14.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 3.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 6.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 25.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 5.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 11.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 9.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 28.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.3 | 4.8 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.3 | 3.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.3 | 5.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 3.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 1.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 13.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.2 | 6.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 7.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 10.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 2.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 6.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 5.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 4.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 3.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 7.3 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 6.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 12.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 2.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 6.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 13.0 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 5.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 2.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 4.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 7.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.6 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 5.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 5.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 9.8 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.1 | 1.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 10.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 3.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 2.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.5 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 4.1 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 2.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 6.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.0 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |