Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
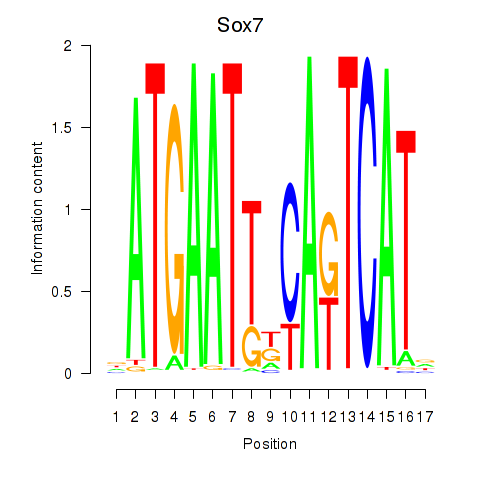
Results for Sox7
Z-value: 0.56
Transcription factors associated with Sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox7
|
ENSMUSG00000063060.5 | SRY (sex determining region Y)-box 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox7 | mm10_v2_chr14_+_63943666_63943687 | -0.31 | 6.5e-02 | Click! |
Activity profile of Sox7 motif
Sorted Z-values of Sox7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_14446570 | 6.53 |
ENSMUST00000063509.4
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr7_-_14446651 | 6.20 |
ENSMUST00000125941.1
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr3_+_146597077 | 2.25 |
ENSMUST00000029837.7
ENSMUST00000121133.1 |
Uox
|
urate oxidase |
| chr7_+_100022099 | 2.23 |
ENSMUST00000144808.1
|
Chrdl2
|
chordin-like 2 |
| chr14_+_66140919 | 2.15 |
ENSMUST00000022620.9
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr11_-_46389509 | 1.76 |
ENSMUST00000020664.6
|
Itk
|
IL2 inducible T cell kinase |
| chr18_+_12741324 | 1.65 |
ENSMUST00000115857.2
ENSMUST00000121018.1 ENSMUST00000119108.1 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr11_-_46389471 | 1.61 |
ENSMUST00000109237.2
|
Itk
|
IL2 inducible T cell kinase |
| chr1_-_136960427 | 1.55 |
ENSMUST00000027649.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_-_146596588 | 1.48 |
ENSMUST00000029836.4
|
Dnase2b
|
deoxyribonuclease II beta |
| chr2_-_120314141 | 1.34 |
ENSMUST00000054651.7
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr9_+_50575273 | 1.25 |
ENSMUST00000059081.6
ENSMUST00000180021.1 |
Il18
|
interleukin 18 |
| chr11_-_46389454 | 1.22 |
ENSMUST00000101306.3
|
Itk
|
IL2 inducible T cell kinase |
| chr19_+_26623419 | 1.13 |
ENSMUST00000176584.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_15055274 | 1.11 |
ENSMUST00000069870.3
|
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr12_-_75735729 | 1.10 |
ENSMUST00000021450.4
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr1_+_140443101 | 0.94 |
ENSMUST00000145077.1
|
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr7_-_48643801 | 0.93 |
ENSMUST00000094383.1
|
Mrgprb3
|
MAS-related GPR, member B3 |
| chr2_+_38511643 | 0.90 |
ENSMUST00000054234.3
ENSMUST00000112902.1 ENSMUST00000112895.1 |
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr7_-_99182681 | 0.83 |
ENSMUST00000033001.4
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr14_+_64652524 | 0.77 |
ENSMUST00000100473.4
|
Kif13b
|
kinesin family member 13B |
| chr3_+_60006743 | 0.75 |
ENSMUST00000169794.1
|
Aadacl2
|
arylacetamide deacetylase-like 2 |
| chr5_-_107875035 | 0.68 |
ENSMUST00000138111.1
ENSMUST00000112642.1 |
Evi5
|
ecotropic viral integration site 5 |
| chr2_-_6130117 | 0.63 |
ENSMUST00000126551.1
ENSMUST00000054254.5 ENSMUST00000114942.2 |
Proser2
|
proline and serine rich 2 |
| chr17_+_34325658 | 0.63 |
ENSMUST00000050325.8
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chrX_-_48513518 | 0.62 |
ENSMUST00000114945.2
ENSMUST00000037349.7 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr1_+_107361929 | 0.57 |
ENSMUST00000027566.2
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr6_+_48684544 | 0.52 |
ENSMUST00000118802.1
|
Gimap4
|
GTPase, IMAP family member 4 |
| chr6_+_48684570 | 0.49 |
ENSMUST00000067506.7
ENSMUST00000119575.1 ENSMUST00000114527.2 ENSMUST00000121957.1 ENSMUST00000090070.5 |
Gimap4
|
GTPase, IMAP family member 4 |
| chrX_-_37168829 | 0.47 |
ENSMUST00000046557.5
|
Akap14
|
A kinase (PRKA) anchor protein 14 |
| chr5_+_8893677 | 0.44 |
ENSMUST00000003717.8
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr10_-_100397072 | 0.43 |
ENSMUST00000167995.1
|
Gm4781
|
predicted gene 4781 |
| chr3_-_19264959 | 0.41 |
ENSMUST00000121951.1
|
Pde7a
|
phosphodiesterase 7A |
| chrX_+_103422010 | 0.38 |
ENSMUST00000182089.1
|
Gm26992
|
predicted gene, 26992 |
| chr6_-_90036841 | 0.37 |
ENSMUST00000073415.1
|
Vmn1r48
|
vomeronasal 1 receptor 48 |
| chr8_+_106935720 | 0.33 |
ENSMUST00000047425.3
|
Sntb2
|
syntrophin, basic 2 |
| chr14_+_53469756 | 0.31 |
ENSMUST00000103643.2
|
Trav8-1
|
T cell receptor alpha variable 8-1 |
| chr7_+_78578830 | 0.27 |
ENSMUST00000064591.4
|
Gm9885
|
predicted gene 9885 |
| chr7_-_7247328 | 0.26 |
ENSMUST00000170922.1
|
Vmn2r29
|
vomeronasal 2, receptor 29 |
| chr2_+_93452796 | 0.24 |
ENSMUST00000099693.2
ENSMUST00000162565.1 ENSMUST00000163052.1 |
Gm10804
|
predicted gene 10804 |
| chr12_-_20900867 | 0.22 |
ENSMUST00000079237.5
|
Zfp125
|
zinc finger protein 125 |
| chr2_+_19344317 | 0.21 |
ENSMUST00000141289.1
|
4930447M23Rik
|
RIKEN cDNA 4930447M23 gene |
| chr16_+_8637674 | 0.20 |
ENSMUST00000023396.9
|
Pmm2
|
phosphomannomutase 2 |
| chr6_-_123533406 | 0.19 |
ENSMUST00000163607.1
|
Vmn2r21
|
vomeronasal 2, receptor 21 |
| chr7_+_45514581 | 0.19 |
ENSMUST00000151506.1
ENSMUST00000085331.5 ENSMUST00000126061.1 |
Tulp2
|
tubby-like protein 2 |
| chr9_+_38773088 | 0.18 |
ENSMUST00000062124.3
|
Olfr921
|
olfactory receptor 921 |
| chr17_+_50239202 | 0.17 |
ENSMUST00000180996.1
|
AY702103
|
cDNA sequence AY702103 |
| chr13_-_4150628 | 0.16 |
ENSMUST00000110704.2
ENSMUST00000021635.7 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr10_-_79614012 | 0.15 |
ENSMUST00000059699.7
ENSMUST00000178228.1 |
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chr9_-_101198999 | 0.14 |
ENSMUST00000066773.7
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr6_-_123418061 | 0.14 |
ENSMUST00000172199.1
|
Vmn2r20
|
vomeronasal 2, receptor 20 |
| chr7_-_8383238 | 0.13 |
ENSMUST00000166499.1
|
Vmn2r44
|
vomeronasal 2, receptor 44 |
| chr2_+_37065746 | 0.12 |
ENSMUST00000120505.1
|
Olfr360
|
olfactory receptor 360 |
| chr2_+_86297817 | 0.11 |
ENSMUST00000054746.2
|
Olfr1052
|
olfactory receptor 1052 |
| chr1_+_169929929 | 0.11 |
ENSMUST00000175731.1
|
1700084C01Rik
|
RIKEN cDNA 1700084C01 gene |
| chr7_+_43609907 | 0.10 |
ENSMUST00000116324.2
|
Zfp819
|
zinc finger protein 819 |
| chr9_-_35648193 | 0.09 |
ENSMUST00000178236.1
|
Gm17365
|
predicted gene, 17365 |
| chr10_-_107912134 | 0.09 |
ENSMUST00000165341.3
|
Otogl
|
otogelin-like |
| chr10_+_31248140 | 0.06 |
ENSMUST00000050717.8
|
Gm5422
|
predicted pseudogene 5422 |
| chr9_+_123767211 | 0.04 |
ENSMUST00000166236.1
ENSMUST00000111454.2 ENSMUST00000168910.1 |
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr2_+_150190393 | 0.04 |
ENSMUST00000109929.2
|
Gm14139
|
predicted gene 14139 |
| chr7_+_16721929 | 0.03 |
ENSMUST00000001984.3
|
Ceacam9
|
carcinoembryonic antigen-related cell adhesion molecule 9 |
| chr7_-_25718976 | 0.01 |
ENSMUST00000002683.2
|
Ccdc97
|
coiled-coil domain containing 97 |
| chr3_-_30140407 | 0.00 |
ENSMUST00000108271.3
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr7_-_7819867 | 0.00 |
ENSMUST00000169683.1
|
Vmn2r35
|
vomeronasal 2, receptor 35 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.3 | 1.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.3 | 4.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 1.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 2.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 2.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.8 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 1.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 2.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.4 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.2 | GO:0009753 | sesquiterpenoid metabolic process(GO:0006714) response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 1.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 2.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.9 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.0 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 4.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 2.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.3 | 0.8 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 1.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 2.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 0.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 4.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 2.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |