Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
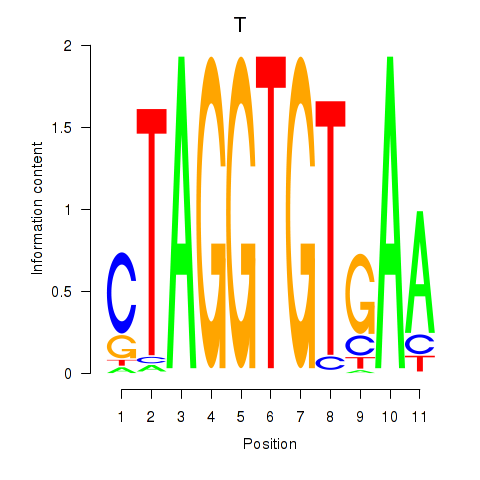
Results for T
Z-value: 0.66
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSMUSG00000062327.4 | brachyury, T-box transcription factor T |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| T | mm10_v2_chr17_+_8434423_8434423 | -0.48 | 3.1e-03 | Click! |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20601958 | 7.35 |
ENSMUST00000087638.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr19_+_37697792 | 5.44 |
ENSMUST00000025946.5
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr9_-_78355743 | 4.46 |
ENSMUST00000125479.1
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr10_+_88459569 | 4.28 |
ENSMUST00000020252.3
ENSMUST00000125612.1 |
Sycp3
|
synaptonemal complex protein 3 |
| chr15_-_60921270 | 4.19 |
ENSMUST00000096418.3
|
A1bg
|
alpha-1-B glycoprotein |
| chr15_+_9335550 | 3.96 |
ENSMUST00000072403.6
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr5_-_87337165 | 3.57 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr19_-_20727533 | 3.56 |
ENSMUST00000025656.3
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr3_-_107943705 | 2.71 |
ENSMUST00000106680.1
ENSMUST00000106684.1 ENSMUST00000106685.2 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr11_-_94392917 | 2.69 |
ENSMUST00000178136.1
ENSMUST00000021231.7 |
Abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr7_+_45259031 | 2.61 |
ENSMUST00000179310.1
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr16_-_18413452 | 2.55 |
ENSMUST00000165430.1
ENSMUST00000147720.1 |
Comt
|
catechol-O-methyltransferase |
| chr1_+_88106834 | 2.45 |
ENSMUST00000113137.1
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr10_-_24101951 | 2.31 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr1_+_88138364 | 2.31 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr3_-_108017877 | 2.09 |
ENSMUST00000004140.4
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr1_-_105356658 | 1.97 |
ENSMUST00000058688.5
ENSMUST00000172299.1 |
Rnf152
|
ring finger protein 152 |
| chr15_-_82407187 | 1.96 |
ENSMUST00000072776.3
|
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr11_-_74925925 | 1.83 |
ENSMUST00000121738.1
|
Srr
|
serine racemase |
| chr19_-_4498574 | 1.80 |
ENSMUST00000048482.6
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr6_+_90333270 | 1.79 |
ENSMUST00000164761.1
ENSMUST00000046128.9 |
Uroc1
|
urocanase domain containing 1 |
| chr5_-_108132513 | 1.73 |
ENSMUST00000119784.1
ENSMUST00000117759.1 |
Tmed5
|
transmembrane emp24 protein transport domain containing 5 |
| chrX_-_139782353 | 1.68 |
ENSMUST00000101217.3
|
Ripply1
|
ripply1 homolog (zebrafish) |
| chr9_+_50494516 | 1.64 |
ENSMUST00000114474.1
|
1600029D21Rik
|
RIKEN cDNA 1600029D21 gene |
| chrX_-_143933089 | 1.63 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr9_+_37613806 | 1.53 |
ENSMUST00000002007.3
|
Siae
|
sialic acid acetylesterase |
| chr2_-_148046896 | 1.49 |
ENSMUST00000172928.1
ENSMUST00000047315.3 |
Foxa2
|
forkhead box A2 |
| chr19_-_28963863 | 1.45 |
ENSMUST00000161813.1
|
4430402I18Rik
|
RIKEN cDNA 4430402I18 gene |
| chr14_-_16243309 | 1.31 |
ENSMUST00000112625.1
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr7_-_72306595 | 1.31 |
ENSMUST00000079323.5
|
Mctp2
|
multiple C2 domains, transmembrane 2 |
| chr5_-_31093500 | 1.30 |
ENSMUST00000031037.7
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr9_-_43239816 | 1.29 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr16_+_93607831 | 1.20 |
ENSMUST00000039659.8
|
Cbr1
|
carbonyl reductase 1 |
| chr8_-_70212251 | 1.15 |
ENSMUST00000063788.7
|
Slc25a42
|
solute carrier family 25, member 42 |
| chrX_+_139610612 | 1.14 |
ENSMUST00000113026.1
|
Rnf128
|
ring finger protein 128 |
| chr2_-_51973219 | 1.07 |
ENSMUST00000028314.2
|
Nmi
|
N-myc (and STAT) interactor |
| chr8_-_70212268 | 1.06 |
ENSMUST00000110127.1
|
Slc25a42
|
solute carrier family 25, member 42 |
| chr15_+_102966794 | 1.06 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chr2_-_181202789 | 1.05 |
ENSMUST00000016511.5
|
Ptk6
|
PTK6 protein tyrosine kinase 6 |
| chr3_+_40950631 | 1.01 |
ENSMUST00000048490.6
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr2_+_160880642 | 1.01 |
ENSMUST00000109456.2
|
Lpin3
|
lipin 3 |
| chr9_+_67759423 | 1.01 |
ENSMUST00000171652.1
|
C2cd4b
|
C2 calcium-dependent domain containing 4B |
| chr4_-_12087912 | 0.99 |
ENSMUST00000050686.3
|
Tmem67
|
transmembrane protein 67 |
| chr13_-_93674300 | 0.99 |
ENSMUST00000015941.7
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr2_-_27248335 | 0.98 |
ENSMUST00000139312.1
|
Sardh
|
sarcosine dehydrogenase |
| chr3_+_90231588 | 0.98 |
ENSMUST00000029546.8
ENSMUST00000119304.1 |
Jtb
|
jumping translocation breakpoint |
| chr12_+_28675220 | 0.95 |
ENSMUST00000020957.6
|
Adi1
|
acireductone dioxygenase 1 |
| chr7_-_74554474 | 0.95 |
ENSMUST00000134539.1
ENSMUST00000026897.7 ENSMUST00000098371.2 |
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr7_-_140882274 | 0.93 |
ENSMUST00000026559.7
|
Sirt3
|
sirtuin 3 |
| chr5_-_38491948 | 0.92 |
ENSMUST00000129099.1
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr4_-_129239165 | 0.86 |
ENSMUST00000097873.3
|
C77080
|
expressed sequence C77080 |
| chr15_-_81360739 | 0.86 |
ENSMUST00000023040.7
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr2_-_51972990 | 0.85 |
ENSMUST00000145481.1
ENSMUST00000112705.2 |
Nmi
|
N-myc (and STAT) interactor |
| chr4_-_129227883 | 0.83 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr11_+_94044331 | 0.81 |
ENSMUST00000024979.8
|
Spag9
|
sperm associated antigen 9 |
| chr2_+_32609043 | 0.79 |
ENSMUST00000128811.1
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr7_-_140881811 | 0.78 |
ENSMUST00000106048.3
ENSMUST00000147331.2 ENSMUST00000137710.1 |
Sirt3
|
sirtuin 3 |
| chrX_-_162565514 | 0.78 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr2_-_103797617 | 0.77 |
ENSMUST00000028607.6
|
Caprin1
|
cell cycle associated protein 1 |
| chr6_-_94700137 | 0.77 |
ENSMUST00000101126.2
ENSMUST00000032105.4 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr4_+_155563700 | 0.77 |
ENSMUST00000105613.3
ENSMUST00000143840.1 ENSMUST00000146080.1 |
Nadk
|
NAD kinase |
| chr6_+_48647224 | 0.75 |
ENSMUST00000078223.3
|
Gimap8
|
GTPase, IMAP family member 8 |
| chr3_-_121283096 | 0.75 |
ENSMUST00000135818.1
ENSMUST00000137234.1 |
Tmem56
|
transmembrane protein 56 |
| chr13_-_81633119 | 0.75 |
ENSMUST00000126444.1
ENSMUST00000128585.1 ENSMUST00000146749.1 ENSMUST00000095585.4 |
Gpr98
|
G protein-coupled receptor 98 |
| chr11_+_94044241 | 0.74 |
ENSMUST00000103168.3
|
Spag9
|
sperm associated antigen 9 |
| chr2_-_80581234 | 0.74 |
ENSMUST00000028386.5
|
Nckap1
|
NCK-associated protein 1 |
| chr7_-_142229971 | 0.74 |
ENSMUST00000097942.2
|
Krtap5-5
|
keratin associated protein 5-5 |
| chr5_+_129895723 | 0.72 |
ENSMUST00000041466.7
ENSMUST00000077320.2 |
2410018M08Rik
|
RIKEN cDNA 2410018M08 gene |
| chr2_+_155133501 | 0.71 |
ENSMUST00000029126.8
ENSMUST00000109685.1 |
Itch
|
itchy, E3 ubiquitin protein ligase |
| chr7_-_114117761 | 0.71 |
ENSMUST00000069449.5
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chrX_-_143933204 | 0.71 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr11_+_94044194 | 0.70 |
ENSMUST00000092777.4
ENSMUST00000075695.6 |
Spag9
|
sperm associated antigen 9 |
| chr16_-_84835484 | 0.70 |
ENSMUST00000114191.1
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr4_+_145696161 | 0.69 |
ENSMUST00000180014.1
|
Gm13242
|
predicted gene 13242 |
| chr7_-_74554726 | 0.69 |
ENSMUST00000107453.1
|
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr2_-_80581380 | 0.68 |
ENSMUST00000111760.2
|
Nckap1
|
NCK-associated protein 1 |
| chr15_+_89386827 | 0.66 |
ENSMUST00000166926.2
|
Klhdc7b
|
kelch domain containing 7B |
| chr14_+_54883377 | 0.65 |
ENSMUST00000022806.3
ENSMUST00000172844.1 ENSMUST00000133397.2 ENSMUST00000134077.1 |
Bcl2l2
Gm20521
|
BCL2-like 2 predicted gene 20521 |
| chr10_+_97693053 | 0.64 |
ENSMUST00000060703.4
|
Ccer1
|
coiled coil glutamate rich protein 1 |
| chr8_+_72492915 | 0.61 |
ENSMUST00000181452.1
|
Gm17435
|
predicted gene, 17435 |
| chr1_-_16619245 | 0.60 |
ENSMUST00000182984.1
ENSMUST00000182554.1 |
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr7_-_79842287 | 0.60 |
ENSMUST00000049004.6
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr11_+_78536355 | 0.60 |
ENSMUST00000128788.1
|
Ift20
|
intraflagellar transport 20 |
| chr4_+_116596672 | 0.59 |
ENSMUST00000051869.7
|
Ccdc17
|
coiled-coil domain containing 17 |
| chr18_+_6765171 | 0.58 |
ENSMUST00000097680.5
|
Rab18
|
RAB18, member RAS oncogene family |
| chr7_-_89517576 | 0.58 |
ENSMUST00000041761.5
|
Prss23
|
protease, serine, 23 |
| chr2_-_126151968 | 0.57 |
ENSMUST00000110448.2
ENSMUST00000110446.2 |
Fam227b
|
family with sequence similarity 227, member B |
| chr11_+_78536393 | 0.57 |
ENSMUST00000050366.8
ENSMUST00000108275.1 |
Ift20
|
intraflagellar transport 20 |
| chr2_+_74681991 | 0.57 |
ENSMUST00000142312.1
|
Hoxd11
|
homeobox D11 |
| chr18_-_12236301 | 0.56 |
ENSMUST00000025279.5
|
Npc1
|
Niemann Pick type C1 |
| chr11_+_79660532 | 0.55 |
ENSMUST00000155381.1
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr9_-_78396096 | 0.53 |
ENSMUST00000171096.1
|
4930542C12Rik
|
RIKEN cDNA 4930542C12 gene |
| chr8_-_105933832 | 0.53 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr7_+_43444104 | 0.53 |
ENSMUST00000004729.3
|
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr2_+_32450444 | 0.52 |
ENSMUST00000048431.2
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr7_-_120095177 | 0.52 |
ENSMUST00000046993.3
|
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr1_-_74749221 | 0.52 |
ENSMUST00000081636.6
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catatlytic subunit |
| chr4_+_53440388 | 0.52 |
ENSMUST00000102911.3
ENSMUST00000107646.2 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr6_-_142008745 | 0.49 |
ENSMUST00000111832.1
|
Gm6614
|
predicted gene 6614 |
| chr7_+_140704165 | 0.49 |
ENSMUST00000080681.3
|
Olfr541
|
olfactory receptor 541 |
| chr17_-_35895920 | 0.48 |
ENSMUST00000059740.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr12_+_84642896 | 0.47 |
ENSMUST00000095551.4
|
Vrtn
|
vertebrae development associated |
| chr7_+_14659496 | 0.47 |
ENSMUST00000183788.1
|
Gm4745
|
predicted gene 4745 |
| chr7_+_96951552 | 0.46 |
ENSMUST00000107159.1
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr18_+_38296805 | 0.46 |
ENSMUST00000171461.1
|
Rnf14
|
ring finger protein 14 |
| chr3_+_109080463 | 0.46 |
ENSMUST00000029478.2
ENSMUST00000159926.1 |
Slc25a54
|
solute carrier family 25, member 54 |
| chr18_+_38296635 | 0.46 |
ENSMUST00000072376.5
ENSMUST00000170811.1 |
Rnf14
|
ring finger protein 14 |
| chr8_-_72492592 | 0.45 |
ENSMUST00000152080.1
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr15_+_88600280 | 0.44 |
ENSMUST00000066949.7
|
Zdhhc25
|
zinc finger, DHHC domain containing 25 |
| chr7_+_14659733 | 0.43 |
ENSMUST00000069740.7
ENSMUST00000183424.1 |
Gm4745
|
predicted gene 4745 |
| chrX_-_162829379 | 0.43 |
ENSMUST00000041370.4
ENSMUST00000112316.2 ENSMUST00000112315.1 |
Txlng
|
taxilin gamma |
| chr11_+_94044111 | 0.42 |
ENSMUST00000132079.1
|
Spag9
|
sperm associated antigen 9 |
| chr8_-_33843562 | 0.42 |
ENSMUST00000183062.1
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr11_+_88047302 | 0.42 |
ENSMUST00000139129.2
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr8_+_70131327 | 0.41 |
ENSMUST00000095273.5
|
Nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr7_-_108930151 | 0.41 |
ENSMUST00000055745.3
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr10_+_128322443 | 0.41 |
ENSMUST00000026446.2
|
Cnpy2
|
canopy 2 homolog (zebrafish) |
| chr1_+_132316112 | 0.40 |
ENSMUST00000082125.5
ENSMUST00000072177.7 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr14_+_53443243 | 0.40 |
ENSMUST00000177622.2
|
Trav7-3
|
T cell receptor alpha variable 7-3 |
| chr11_+_60417238 | 0.39 |
ENSMUST00000070681.6
|
Gid4
|
GID complex subunit 4, VID24 homolog (S. cerevisiae) |
| chr7_-_15627876 | 0.38 |
ENSMUST00000086122.3
ENSMUST00000174443.1 |
Obox3
|
oocyte specific homeobox 3 |
| chr8_+_106935720 | 0.38 |
ENSMUST00000047425.3
|
Sntb2
|
syntrophin, basic 2 |
| chr1_-_56971762 | 0.38 |
ENSMUST00000114415.3
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr14_+_31019125 | 0.36 |
ENSMUST00000112095.1
ENSMUST00000112098.3 ENSMUST00000112106.1 ENSMUST00000146325.1 |
Pbrm1
|
polybromo 1 |
| chr8_+_113643206 | 0.36 |
ENSMUST00000034219.4
ENSMUST00000095173.1 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr16_-_10796815 | 0.32 |
ENSMUST00000023144.5
|
Prm1
|
protamine 1 |
| chr7_+_15750377 | 0.32 |
ENSMUST00000098802.3
ENSMUST00000173053.1 |
Obox5
|
oocyte specific homeobox 5 |
| chr1_-_156032948 | 0.30 |
ENSMUST00000136397.1
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr10_+_99420522 | 0.27 |
ENSMUST00000060657.2
|
B530045E10Rik
|
RIKEN cDNA B530045E10 gene |
| chr2_+_74697663 | 0.27 |
ENSMUST00000059272.8
|
Hoxd9
|
homeobox D9 |
| chr9_-_31464238 | 0.27 |
ENSMUST00000048050.7
|
Tmem45b
|
transmembrane protein 45b |
| chr7_-_30635852 | 0.26 |
ENSMUST00000108147.2
|
Etv2
|
ets variant gene 2 |
| chr17_+_46496753 | 0.26 |
ENSMUST00000046497.6
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr16_-_44333135 | 0.26 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr16_-_44332925 | 0.25 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr14_-_100149764 | 0.25 |
ENSMUST00000097079.4
|
Klf12
|
Kruppel-like factor 12 |
| chr7_-_79793788 | 0.25 |
ENSMUST00000032760.5
|
Mesp1
|
mesoderm posterior 1 |
| chr6_-_140041409 | 0.24 |
ENSMUST00000032356.6
|
Plcz1
|
phospholipase C, zeta 1 |
| chr3_+_152346465 | 0.24 |
ENSMUST00000026507.6
ENSMUST00000117492.2 |
Usp33
|
ubiquitin specific peptidase 33 |
| chr12_-_111966954 | 0.23 |
ENSMUST00000021719.5
|
2010107E04Rik
|
RIKEN cDNA 2010107E04 gene |
| chr11_-_98193260 | 0.23 |
ENSMUST00000092735.5
ENSMUST00000107545.2 |
Med1
|
mediator complex subunit 1 |
| chr1_-_156421146 | 0.23 |
ENSMUST00000178036.1
|
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr10_+_21978643 | 0.22 |
ENSMUST00000142174.1
ENSMUST00000164659.1 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_-_101893789 | 0.22 |
ENSMUST00000051043.3
|
C130073F10Rik
|
RIKEN cDNA C130073F10 gene |
| chr16_-_44746337 | 0.21 |
ENSMUST00000023348.4
ENSMUST00000162512.1 |
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr4_-_3574844 | 0.21 |
ENSMUST00000029891.5
|
Tmem68
|
transmembrane protein 68 |
| chr9_+_13765970 | 0.21 |
ENSMUST00000152532.1
|
Mtmr2
|
myotubularin related protein 2 |
| chr16_+_84835070 | 0.21 |
ENSMUST00000009120.7
|
Gabpa
|
GA repeat binding protein, alpha |
| chr1_-_16619437 | 0.20 |
ENSMUST00000117146.1
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr6_-_24956106 | 0.20 |
ENSMUST00000127247.2
|
Tmem229a
|
transmembrane protein 229A |
| chrY_+_897782 | 0.20 |
ENSMUST00000055032.7
|
Kdm5d
|
lysine (K)-specific demethylase 5D |
| chr5_+_22775630 | 0.19 |
ENSMUST00000179257.1
|
Gm21846
|
predicted gene, 21846 |
| chr14_+_3892557 | 0.19 |
ENSMUST00000178392.1
|
Gm2916
|
predicted gene 2916 |
| chr2_+_126152141 | 0.19 |
ENSMUST00000170908.1
|
Dtwd1
|
DTW domain containing 1 |
| chr10_-_103236280 | 0.19 |
ENSMUST00000123364.1
ENSMUST00000166240.1 ENSMUST00000020043.5 |
Lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr11_+_105178765 | 0.18 |
ENSMUST00000106939.2
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr9_-_110989611 | 0.18 |
ENSMUST00000084922.5
|
Rtp3
|
receptor transporter protein 3 |
| chr12_+_84316830 | 0.17 |
ENSMUST00000045931.10
|
Zfp410
|
zinc finger protein 410 |
| chr11_-_86544754 | 0.17 |
ENSMUST00000138810.1
ENSMUST00000058286.2 ENSMUST00000154617.1 |
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1 |
| chr11_+_88047788 | 0.16 |
ENSMUST00000107920.3
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr7_-_107757993 | 0.16 |
ENSMUST00000052438.7
|
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr14_+_41570696 | 0.16 |
ENSMUST00000100701.4
|
1700049E17Rik2
|
tandem duplication of RIKEN cDNA 1700049E17 gene, gene 2 |
| chr11_-_65159890 | 0.15 |
ENSMUST00000020855.3
ENSMUST00000108696.1 |
1700086D15Rik
|
RIKEN cDNA 1700086D15 gene |
| chr1_-_44218952 | 0.15 |
ENSMUST00000054801.3
|
Mettl21e
|
methyltransferase like 21E |
| chr11_-_78536235 | 0.15 |
ENSMUST00000017759.2
ENSMUST00000108277.2 |
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr13_-_38037069 | 0.14 |
ENSMUST00000089840.4
|
Cage1
|
cancer antigen 1 |
| chr14_-_41069074 | 0.13 |
ENSMUST00000022316.4
|
Dydc2
|
DPY30 domain containing 2 |
| chr15_-_100403243 | 0.13 |
ENSMUST00000124324.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr8_-_15046047 | 0.13 |
ENSMUST00000050493.3
ENSMUST00000123331.1 |
BB014433
|
expressed sequence BB014433 |
| chr11_-_60417022 | 0.12 |
ENSMUST00000108721.1
|
Atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr5_-_142906702 | 0.12 |
ENSMUST00000167721.1
ENSMUST00000163829.1 ENSMUST00000100497.4 |
Actb
|
actin, beta |
| chr4_+_155624853 | 0.12 |
ENSMUST00000067081.3
ENSMUST00000105600.1 ENSMUST00000105598.1 |
Cdk11b
|
cyclin-dependent kinase 11B |
| chr7_+_15750134 | 0.11 |
ENSMUST00000173455.1
|
Obox5
|
oocyte specific homeobox 5 |
| chr2_-_30178422 | 0.10 |
ENSMUST00000100220.4
ENSMUST00000179795.1 |
D2Wsu81e
|
DNA segment, Chr 2, Wayne State University 81, expressed |
| chr14_-_21740395 | 0.10 |
ENSMUST00000120984.2
|
Dusp13
|
dual specificity phosphatase 13 |
| chr1_+_97770158 | 0.09 |
ENSMUST00000112844.3
ENSMUST00000112842.1 ENSMUST00000027571.6 |
Gin1
|
gypsy retrotransposon integrase 1 |
| chr14_+_31019159 | 0.08 |
ENSMUST00000112094.1
ENSMUST00000144009.1 |
Pbrm1
|
polybromo 1 |
| chr6_+_52714219 | 0.08 |
ENSMUST00000138040.1
ENSMUST00000129660.1 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr19_+_23758819 | 0.06 |
ENSMUST00000025830.7
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr18_-_36726730 | 0.05 |
ENSMUST00000061829.6
|
Cd14
|
CD14 antigen |
| chr1_-_71414881 | 0.04 |
ENSMUST00000087268.5
|
Abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr11_-_65269984 | 0.04 |
ENSMUST00000108695.2
|
Myocd
|
myocardin |
| chr10_-_116896879 | 0.04 |
ENSMUST00000048229.7
|
Myrfl
|
myelin regulatory factor-like |
| chr6_-_116716888 | 0.03 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr4_-_41731142 | 0.02 |
ENSMUST00000171251.1
ENSMUST00000159930.1 |
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr15_-_59374149 | 0.02 |
ENSMUST00000022976.4
|
E430025E21Rik
|
RIKEN cDNA E430025E21 gene |
| chr4_-_133277730 | 0.02 |
ENSMUST00000105907.2
|
Tmem222
|
transmembrane protein 222 |
| chr5_+_73406076 | 0.01 |
ENSMUST00000031040.6
ENSMUST00000065543.6 |
Cwh43
|
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) |
| chrX_-_111536325 | 0.00 |
ENSMUST00000156639.1
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr11_-_69666062 | 0.00 |
ENSMUST00000108654.2
ENSMUST00000018918.5 |
Cd68
|
CD68 antigen |
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.1 | 4.3 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) female meiosis sister chromatid cohesion(GO:0007066) |
| 1.0 | 7.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.9 | 2.6 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.7 | 2.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.6 | 1.9 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.5 | 2.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.5 | 1.5 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.5 | 8.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.4 | 1.8 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.4 | 1.8 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.4 | 3.6 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 2.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 1.0 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.3 | 1.9 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.3 | 8.4 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.3 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 1.3 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 0.9 | GO:0015867 | ATP transport(GO:0015867) |
| 0.2 | 1.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 0.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 1.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 2.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.3 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 2.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 2.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.3 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.6 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.4 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.8 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.4 | GO:0007128 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.0 | 1.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.9 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.1 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0071725 | response to diacyl bacterial lipopeptide(GO:0071724) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to diacyl bacterial lipopeptide(GO:0071726) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.8 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 1.7 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.2 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.5 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0000802 | transverse filament(GO:0000802) |
| 0.3 | 1.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.5 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 1.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 3.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 6.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.8 | 5.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.0 | 3.1 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.9 | 2.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.6 | 1.8 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.4 | 2.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.3 | 1.0 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.3 | 1.3 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 12.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 1.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 1.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 2.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 6.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 2.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 1.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 0.5 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 1.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 2.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 3.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 2.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 5.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 6.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |