Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tal1
Z-value: 4.01
Transcription factors associated with Tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tal1
|
ENSMUSG00000028717.6 | T cell acute lymphocytic leukemia 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tal1 | mm10_v2_chr4_+_115059507_115059553 | 0.97 | 3.0e-22 | Click! |
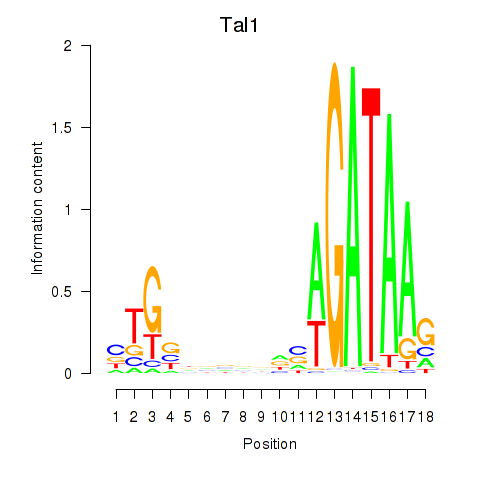
Activity profile of Tal1 motif
Sorted Z-values of Tal1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_132367879 | 62.65 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr14_-_43875517 | 60.38 |
ENSMUST00000179200.1
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr6_+_86078070 | 58.34 |
ENSMUST00000032069.5
|
Add2
|
adducin 2 (beta) |
| chr4_-_119189949 | 49.25 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr15_-_103251465 | 49.22 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr17_+_40811089 | 48.77 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr7_-_103853199 | 48.10 |
ENSMUST00000033229.3
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr4_+_134864536 | 40.10 |
ENSMUST00000030627.7
|
Rhd
|
Rh blood group, D antigen |
| chr14_-_43819639 | 39.84 |
ENSMUST00000100691.3
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr1_+_174172738 | 38.09 |
ENSMUST00000027817.7
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr11_+_32276400 | 37.79 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr16_-_18621366 | 36.05 |
ENSMUST00000051160.2
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr17_+_36869567 | 35.63 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr14_+_51853699 | 35.26 |
ENSMUST00000169070.1
ENSMUST00000074477.6 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chr1_+_131638485 | 34.73 |
ENSMUST00000112411.1
|
Ctse
|
cathepsin E |
| chr4_-_119190005 | 33.95 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr17_-_26199008 | 33.88 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr11_+_32276893 | 33.12 |
ENSMUST00000145569.1
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr14_+_27000362 | 33.07 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr1_+_131638306 | 32.86 |
ENSMUST00000073350.6
|
Ctse
|
cathepsin E |
| chrX_+_8271642 | 32.15 |
ENSMUST00000115590.1
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr7_-_4397705 | 31.80 |
ENSMUST00000108590.2
|
Gp6
|
glycoprotein 6 (platelet) |
| chr15_+_80623499 | 30.52 |
ENSMUST00000043149.7
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr17_+_31208049 | 30.29 |
ENSMUST00000173776.1
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr8_+_80494032 | 29.62 |
ENSMUST00000063359.6
|
Gypa
|
glycophorin A |
| chr8_+_84701430 | 29.16 |
ENSMUST00000037165.4
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr7_+_110773658 | 27.82 |
ENSMUST00000143786.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr11_+_58918004 | 27.33 |
ENSMUST00000108818.3
ENSMUST00000020792.5 |
Btnl10
|
butyrophilin-like 10 |
| chr8_-_85365317 | 25.48 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_85365341 | 25.34 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr17_-_28560704 | 24.65 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chrX_+_93675088 | 24.27 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr9_-_21963568 | 23.79 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr11_+_58917889 | 22.99 |
ENSMUST00000069941.6
|
Btnl10
|
butyrophilin-like 10 |
| chr10_-_79788924 | 22.63 |
ENSMUST00000020573.6
|
Prss57
|
protease, serine 57 |
| chr7_-_100467149 | 22.55 |
ENSMUST00000184420.1
|
RP23-308M1.2
|
RP23-308M1.2 |
| chr7_+_99594605 | 22.22 |
ENSMUST00000162290.1
|
Arrb1
|
arrestin, beta 1 |
| chr9_+_21029373 | 22.04 |
ENSMUST00000001040.5
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr2_+_129228022 | 21.41 |
ENSMUST00000148548.1
|
A730036I17Rik
|
RIKEN cDNA A730036I17 gene |
| chr13_+_108316395 | 21.06 |
ENSMUST00000171178.1
|
Depdc1b
|
DEP domain containing 1B |
| chr2_+_84988194 | 20.88 |
ENSMUST00000028466.5
|
Prg3
|
proteoglycan 3 |
| chr10_-_62342674 | 19.80 |
ENSMUST00000143179.1
ENSMUST00000130422.1 |
Hk1
|
hexokinase 1 |
| chr6_+_41354105 | 19.63 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr7_-_99238564 | 19.10 |
ENSMUST00000064231.7
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr5_+_115845229 | 18.88 |
ENSMUST00000137952.1
ENSMUST00000148245.1 |
Cit
|
citron |
| chr6_+_41458923 | 18.76 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr11_-_80080928 | 17.65 |
ENSMUST00000103233.3
ENSMUST00000061283.8 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr6_-_41314700 | 17.04 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr10_-_80421847 | 16.63 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chr19_+_58759700 | 15.86 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr11_+_70639118 | 15.67 |
ENSMUST00000055184.6
ENSMUST00000108551.2 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr7_+_43351378 | 15.25 |
ENSMUST00000012798.7
ENSMUST00000122423.1 ENSMUST00000121494.1 |
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr3_-_98893209 | 14.56 |
ENSMUST00000029464.7
|
Hao2
|
hydroxyacid oxidase 2 |
| chr17_+_34914459 | 14.40 |
ENSMUST00000007249.8
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr5_-_24030297 | 14.33 |
ENSMUST00000101513.2
|
Fam126a
|
family with sequence similarity 126, member A |
| chr7_+_142498832 | 14.11 |
ENSMUST00000078497.8
ENSMUST00000105953.3 ENSMUST00000179658.1 ENSMUST00000105954.3 ENSMUST00000105952.3 ENSMUST00000105955.1 ENSMUST00000074187.6 ENSMUST00000180152.1 ENSMUST00000105950.4 ENSMUST00000105957.3 ENSMUST00000169299.2 ENSMUST00000105958.3 ENSMUST00000105949.1 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr6_+_30541582 | 13.57 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr11_+_58948890 | 13.50 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chrX_-_107403295 | 13.48 |
ENSMUST00000033591.5
|
Itm2a
|
integral membrane protein 2A |
| chr3_+_103832562 | 13.14 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr15_+_57985873 | 12.80 |
ENSMUST00000050374.2
|
Fam83a
|
family with sequence similarity 83, member A |
| chr9_+_107950952 | 12.72 |
ENSMUST00000049348.3
|
Traip
|
TRAF-interacting protein |
| chr2_+_84839395 | 12.43 |
ENSMUST00000146816.1
ENSMUST00000028469.7 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr4_-_41464816 | 12.05 |
ENSMUST00000108055.2
ENSMUST00000154535.1 ENSMUST00000030148.5 |
Kif24
|
kinesin family member 24 |
| chr4_-_42874195 | 12.04 |
ENSMUST00000107978.1
ENSMUST00000055944.4 |
BC049635
|
cDNA sequence BC049635 |
| chr1_-_173333503 | 11.90 |
ENSMUST00000038227.4
|
Darc
|
Duffy blood group, chemokine receptor |
| chr11_+_117782358 | 11.69 |
ENSMUST00000117781.1
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr2_-_153241402 | 11.43 |
ENSMUST00000056924.7
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr11_+_117782076 | 11.36 |
ENSMUST00000127080.1
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr11_+_117782281 | 11.21 |
ENSMUST00000050874.7
ENSMUST00000106334.2 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr6_-_136857727 | 10.44 |
ENSMUST00000032341.2
|
Art4
|
ADP-ribosyltransferase 4 |
| chr4_+_132974102 | 10.37 |
ENSMUST00000030693.6
|
Fgr
|
Gardner-Rasheed feline sarcoma viral (Fgr) oncogene homolog |
| chr13_-_95525239 | 10.33 |
ENSMUST00000022185.8
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr11_-_102469839 | 10.07 |
ENSMUST00000103086.3
|
Itga2b
|
integrin alpha 2b |
| chr2_-_153225396 | 9.51 |
ENSMUST00000099194.2
|
Tspyl3
|
TSPY-like 3 |
| chr4_-_154928187 | 9.32 |
ENSMUST00000123514.1
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr9_-_111057235 | 9.17 |
ENSMUST00000111888.1
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr8_-_77517898 | 8.86 |
ENSMUST00000076316.4
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr17_-_29237759 | 8.82 |
ENSMUST00000137727.1
ENSMUST00000024805.7 |
Cpne5
|
copine V |
| chr14_+_44102654 | 8.64 |
ENSMUST00000074839.6
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr15_-_34356421 | 8.57 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr11_-_117782182 | 8.52 |
ENSMUST00000152304.1
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr5_-_24030649 | 8.38 |
ENSMUST00000030849.6
|
Fam126a
|
family with sequence similarity 126, member A |
| chr4_+_155790439 | 8.18 |
ENSMUST00000165000.1
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr14_-_43923559 | 8.10 |
ENSMUST00000159175.1
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr1_-_171649002 | 8.02 |
ENSMUST00000111276.3
|
Slamf7
|
SLAM family member 7 |
| chr1_-_52232296 | 7.90 |
ENSMUST00000114512.1
|
Gls
|
glutaminase |
| chr17_-_33713372 | 7.90 |
ENSMUST00000173392.1
|
March2
|
membrane-associated ring finger (C3HC4) 2 |
| chr14_-_43923368 | 7.86 |
ENSMUST00000163652.1
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr1_-_181842334 | 7.65 |
ENSMUST00000005003.6
|
Lbr
|
lamin B receptor |
| chr5_+_138280538 | 7.58 |
ENSMUST00000162245.1
ENSMUST00000161691.1 |
Stag3
|
stromal antigen 3 |
| chr5_+_115466234 | 7.48 |
ENSMUST00000145785.1
ENSMUST00000031495.4 ENSMUST00000112071.1 ENSMUST00000125568.1 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr5_+_138280516 | 7.41 |
ENSMUST00000048028.8
|
Stag3
|
stromal antigen 3 |
| chr3_-_14808358 | 7.28 |
ENSMUST00000181860.1
ENSMUST00000144327.2 |
Car1
|
carbonic anhydrase 1 |
| chr6_+_125071277 | 7.27 |
ENSMUST00000140346.2
ENSMUST00000171989.1 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr7_+_97842917 | 6.99 |
ENSMUST00000033040.5
|
Pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr6_-_25809189 | 6.81 |
ENSMUST00000115327.1
|
Pot1a
|
protection of telomeres 1A |
| chr6_-_25809210 | 6.81 |
ENSMUST00000115330.1
ENSMUST00000115329.1 |
Pot1a
|
protection of telomeres 1A |
| chr7_-_81566939 | 6.76 |
ENSMUST00000042318.5
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr2_+_5137756 | 6.59 |
ENSMUST00000027988.7
|
Ccdc3
|
coiled-coil domain containing 3 |
| chr16_-_19983005 | 6.55 |
ENSMUST00000058839.8
|
Klhl6
|
kelch-like 6 |
| chr3_+_79884576 | 6.41 |
ENSMUST00000145992.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr7_+_103937382 | 6.41 |
ENSMUST00000098189.1
|
Olfr632
|
olfactory receptor 632 |
| chr15_+_78926720 | 6.40 |
ENSMUST00000089377.5
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr9_-_42399709 | 6.33 |
ENSMUST00000160940.1
|
Tecta
|
tectorin alpha |
| chr10_+_97565436 | 6.33 |
ENSMUST00000038160.4
|
Lum
|
lumican |
| chr11_+_70505244 | 6.23 |
ENSMUST00000019063.2
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr8_+_123117354 | 5.96 |
ENSMUST00000037900.8
|
Cpne7
|
copine VII |
| chrX_+_56447965 | 5.89 |
ENSMUST00000079663.6
|
Gm2174
|
predicted gene 2174 |
| chrX_-_139085211 | 5.77 |
ENSMUST00000033626.8
ENSMUST00000060824.3 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr11_+_4986824 | 5.71 |
ENSMUST00000009234.9
ENSMUST00000109897.1 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr19_-_29367294 | 5.50 |
ENSMUST00000138051.1
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr1_+_171559186 | 5.47 |
ENSMUST00000004829.7
|
Cd244
|
CD244 natural killer cell receptor 2B4 |
| chr14_+_33954020 | 5.38 |
ENSMUST00000035695.8
|
Rbp3
|
retinol binding protein 3, interstitial |
| chr5_-_24351604 | 5.19 |
ENSMUST00000036092.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr14_-_78725089 | 5.17 |
ENSMUST00000074729.5
|
Dgkh
|
diacylglycerol kinase, eta |
| chr5_-_137116177 | 4.94 |
ENSMUST00000054384.5
ENSMUST00000152207.1 |
Trim56
|
tripartite motif-containing 56 |
| chrX_-_139085230 | 4.93 |
ENSMUST00000152457.1
|
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr11_-_78176619 | 4.91 |
ENSMUST00000148154.2
ENSMUST00000017549.6 |
Nek8
|
NIMA (never in mitosis gene a)-related expressed kinase 8 |
| chr13_+_49682191 | 4.90 |
ENSMUST00000172254.1
|
Iars
|
isoleucine-tRNA synthetase |
| chr5_+_137630116 | 4.84 |
ENSMUST00000175968.1
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr2_-_102901346 | 4.83 |
ENSMUST00000111192.2
ENSMUST00000111190.2 ENSMUST00000111198.2 ENSMUST00000111191.2 ENSMUST00000060516.7 ENSMUST00000099673.2 ENSMUST00000005218.8 ENSMUST00000111194.1 |
Cd44
|
CD44 antigen |
| chr10_+_99443699 | 4.79 |
ENSMUST00000167243.1
|
Gad1-ps
|
glutamate decarboxylase 1, pseudogene |
| chr10_-_93891141 | 4.66 |
ENSMUST00000180840.1
|
Metap2
|
methionine aminopeptidase 2 |
| chr4_-_56802265 | 4.54 |
ENSMUST00000030140.2
|
Ikbkap
|
inhibitor of kappa light polypeptide enhancer in B cells, kinase complex-associated protein |
| chr14_+_51129055 | 4.42 |
ENSMUST00000095923.3
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr15_-_74763567 | 4.32 |
ENSMUST00000040404.6
|
Ly6d
|
lymphocyte antigen 6 complex, locus D |
| chr4_-_32923455 | 4.25 |
ENSMUST00000035719.4
ENSMUST00000084749.1 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr17_+_27342453 | 4.19 |
ENSMUST00000151398.1
|
Gm10505
|
predicted gene 10505 |
| chr19_+_29367447 | 4.09 |
ENSMUST00000016640.7
|
Cd274
|
CD274 antigen |
| chr10_-_129902726 | 3.90 |
ENSMUST00000071557.1
|
Olfr815
|
olfactory receptor 815 |
| chr1_-_173912904 | 3.88 |
ENSMUST00000009340.8
|
Mnda
|
myeloid cell nuclear differentiation antigen |
| chr11_+_67200052 | 3.82 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr4_+_56802337 | 3.79 |
ENSMUST00000045368.5
|
BC026590
|
cDNA sequence BC026590 |
| chr11_+_94936224 | 3.79 |
ENSMUST00000001547.7
|
Col1a1
|
collagen, type I, alpha 1 |
| chr6_+_29279587 | 3.76 |
ENSMUST00000167131.1
|
Fam71f2
|
family with sequence similarity 71, member F2 |
| chr1_-_173535957 | 3.74 |
ENSMUST00000139092.1
|
BC094916
|
cDNA sequence BC094916 |
| chr4_+_132535542 | 3.67 |
ENSMUST00000094657.3
ENSMUST00000105940.3 ENSMUST00000105939.3 ENSMUST00000150207.1 |
Dnajc8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr17_-_7385305 | 3.66 |
ENSMUST00000070059.3
|
Gm9992
|
predicted gene 9992 |
| chr1_-_167466780 | 3.60 |
ENSMUST00000036643.4
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr7_-_15999495 | 3.58 |
ENSMUST00000094821.3
|
Gltscr1
|
glioma tumor suppressor candidate region gene 1 |
| chr10_+_116177351 | 3.55 |
ENSMUST00000155606.1
ENSMUST00000128399.1 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr3_+_98222148 | 3.52 |
ENSMUST00000029469.4
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr15_+_12205009 | 3.49 |
ENSMUST00000038172.8
|
Mtmr12
|
myotubularin related protein 12 |
| chr3_+_79884496 | 3.47 |
ENSMUST00000118853.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr11_-_98625661 | 3.36 |
ENSMUST00000104933.1
|
Gm12355
|
predicted gene 12355 |
| chr2_-_120731503 | 3.36 |
ENSMUST00000110701.1
ENSMUST00000110700.1 |
Cdan1
|
congenital dyserythropoietic anemia, type I (human) |
| chr14_+_76504185 | 3.32 |
ENSMUST00000177207.1
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr11_+_50850685 | 3.18 |
ENSMUST00000171427.1
|
Grm6
|
glutamate receptor, metabotropic 6 |
| chr9_-_32541589 | 3.16 |
ENSMUST00000016231.7
|
Fli1
|
Friend leukemia integration 1 |
| chr9_+_66350465 | 3.10 |
ENSMUST00000042824.6
|
Herc1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| chr6_+_41302265 | 3.09 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr2_+_180499893 | 3.05 |
ENSMUST00000029084.2
|
Ntsr1
|
neurotensin receptor 1 |
| chr19_-_46969474 | 2.93 |
ENSMUST00000086961.7
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr10_+_39133981 | 2.89 |
ENSMUST00000019991.7
|
Tube1
|
epsilon-tubulin 1 |
| chr3_+_88621436 | 2.89 |
ENSMUST00000170653.2
ENSMUST00000177303.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr10_-_128626464 | 2.80 |
ENSMUST00000026420.5
|
Rps26
|
ribosomal protein S26 |
| chr14_+_22019833 | 2.76 |
ENSMUST00000159777.1
ENSMUST00000162540.1 |
1700112E06Rik
|
RIKEN cDNA 1700112E06 gene |
| chr1_-_160792908 | 2.67 |
ENSMUST00000028049.7
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr14_+_22019712 | 2.66 |
ENSMUST00000075639.4
ENSMUST00000161249.1 |
1700112E06Rik
|
RIKEN cDNA 1700112E06 gene |
| chr2_+_136891501 | 2.65 |
ENSMUST00000141463.1
|
Slx4ip
|
SLX4 interacting protein |
| chr6_+_47877204 | 2.64 |
ENSMUST00000061890.7
|
Zfp282
|
zinc finger protein 282 |
| chr13_-_13393592 | 2.63 |
ENSMUST00000021738.8
|
Gpr137b
|
G protein-coupled receptor 137B |
| chr11_-_99322943 | 2.60 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr10_+_41887428 | 2.59 |
ENSMUST00000041438.6
|
Sesn1
|
sestrin 1 |
| chr2_+_164746028 | 2.50 |
ENSMUST00000109327.3
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr7_-_18992774 | 2.43 |
ENSMUST00000098778.2
|
Gm10676
|
predicted gene 10676 |
| chrX_+_73757069 | 2.41 |
ENSMUST00000002079.6
|
Plxnb3
|
plexin B3 |
| chr4_-_118179946 | 2.29 |
ENSMUST00000050288.8
ENSMUST00000106403.1 |
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr6_-_34317442 | 2.12 |
ENSMUST00000154655.1
ENSMUST00000102980.4 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr9_-_62510498 | 2.00 |
ENSMUST00000164246.2
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr17_+_16972910 | 1.92 |
ENSMUST00000071374.5
|
BC002059
|
cDNA sequence BC002059 |
| chrX_-_134111852 | 1.92 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr10_+_116177217 | 1.83 |
ENSMUST00000148731.1
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr13_+_107031963 | 1.79 |
ENSMUST00000095459.1
|
3830408C21Rik
|
RIKEN cDNA 3830408C21 gene |
| chr2_+_164745979 | 1.78 |
ENSMUST00000017443.7
ENSMUST00000109326.3 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr2_-_6721606 | 1.68 |
ENSMUST00000150624.2
ENSMUST00000142941.1 ENSMUST00000100429.4 ENSMUST00000182879.1 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr7_-_135528645 | 1.68 |
ENSMUST00000053716.7
|
Clrn3
|
clarin 3 |
| chr5_+_29378604 | 1.66 |
ENSMUST00000181005.1
|
4632411P08Rik
|
RIKEN cDNA 4632411P08 gene |
| chr14_+_51162260 | 1.59 |
ENSMUST00000075648.3
|
Ear5
|
eosinophil-associated, ribonuclease A family, member 5 |
| chr12_+_95692212 | 1.59 |
ENSMUST00000057324.3
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_134906726 | 1.56 |
ENSMUST00000047305.5
|
Wbscr28
|
Williams-Beuren syndrome chromosome region 28 (human) |
| chr9_-_60688118 | 1.55 |
ENSMUST00000114034.2
ENSMUST00000065603.5 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr2_+_127080252 | 1.55 |
ENSMUST00000142737.1
|
Blvra
|
biliverdin reductase A |
| chr17_+_34354787 | 1.48 |
ENSMUST00000178562.1
ENSMUST00000025198.7 |
Btnl2
|
butyrophilin-like 2 |
| chr5_-_5265224 | 1.37 |
ENSMUST00000115450.1
|
Cdk14
|
cyclin-dependent kinase 14 |
| chr2_+_136892168 | 1.35 |
ENSMUST00000099311.2
|
Slx4ip
|
SLX4 interacting protein |
| chr10_-_117224480 | 1.34 |
ENSMUST00000020382.6
|
Yeats4
|
YEATS domain containing 4 |
| chr7_+_16875302 | 1.26 |
ENSMUST00000108493.1
|
Dact3
|
dapper homolog 3, antagonist of beta-catenin (xenopus) |
| chr15_+_85017138 | 1.23 |
ENSMUST00000023070.5
|
Upk3a
|
uroplakin 3A |
| chrX_-_134111708 | 1.12 |
ENSMUST00000159259.1
ENSMUST00000113275.3 |
Nox1
|
NADPH oxidase 1 |
| chr4_-_59438633 | 1.12 |
ENSMUST00000040166.7
ENSMUST00000107544.1 |
Susd1
|
sushi domain containing 1 |
| chr2_+_30061754 | 1.07 |
ENSMUST00000149578.1
ENSMUST00000102866.3 |
Set
|
SET nuclear oncogene |
| chr7_-_45896656 | 1.03 |
ENSMUST00000120299.1
|
Syngr4
|
synaptogyrin 4 |
| chr10_+_33863935 | 1.00 |
ENSMUST00000092597.3
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr4_-_43669141 | 0.97 |
ENSMUST00000056474.6
|
Fam221b
|
family with sequence similarity 221, member B |
| chr2_-_6721890 | 0.83 |
ENSMUST00000114927.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr2_+_144033059 | 0.77 |
ENSMUST00000037722.2
ENSMUST00000110032.1 |
Banf2
|
barrier to autointegration factor 2 |
| chrX_+_144153695 | 0.77 |
ENSMUST00000135687.1
|
A730046J19Rik
|
RIKEN cDNA A730046J19 gene |
| chr11_+_65807175 | 0.75 |
ENSMUST00000071465.2
ENSMUST00000018491.7 |
Zkscan6
|
zinc finger with KRAB and SCAN domains 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tal1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.6 | 88.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 14.9 | 119.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 8.5 | 50.8 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 6.4 | 19.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 5.2 | 20.9 | GO:0045575 | basophil activation(GO:0045575) |
| 4.8 | 14.4 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 4.5 | 13.6 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 4.2 | 67.6 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 4.2 | 29.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 4.1 | 49.2 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 3.7 | 15.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 3.7 | 22.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 3.7 | 29.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 3.6 | 10.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 3.4 | 10.3 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 3.3 | 19.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 3.3 | 33.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 3.1 | 27.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 2.9 | 65.9 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 2.5 | 7.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 2.5 | 19.8 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 2.1 | 58.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 2.1 | 16.6 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 1.9 | 5.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 1.8 | 5.5 | GO:0071661 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 1.8 | 32.2 | GO:0015816 | glycine transport(GO:0015816) |
| 1.8 | 40.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 1.6 | 23.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 1.4 | 14.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 1.4 | 7.0 | GO:0060244 | amygdala development(GO:0021764) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.3 | 18.7 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 1.3 | 15.9 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 1.3 | 34.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 1.3 | 7.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.3 | 38.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 1.3 | 5.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 1.0 | 62.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 1.0 | 36.9 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.9 | 5.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.9 | 18.9 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.7 | 2.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.7 | 22.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.7 | 15.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.6 | 5.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.6 | 2.4 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.6 | 4.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.6 | 4.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.6 | 14.6 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.5 | 10.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.5 | 3.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.5 | 12.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.5 | 1.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.4 | 4.9 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.4 | 4.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.4 | 18.1 | GO:0032094 | response to food(GO:0032094) |
| 0.4 | 8.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.4 | 4.7 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.4 | 7.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.4 | 2.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 27.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.4 | 4.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.4 | 24.3 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.3 | 5.2 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.3 | 4.8 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 5.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 2.9 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.3 | 6.5 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.3 | 5.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 0.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 10.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 3.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 | 45.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.2 | 8.9 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.2 | 11.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 11.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 1.5 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 12.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 2.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 1.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.2 | 21.1 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.2 | 3.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 7.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 19.1 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 4.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 17.9 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.1 | 3.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 6.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 8.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 2.6 | GO:0045109 | hair follicle morphogenesis(GO:0031069) intermediate filament organization(GO:0045109) |
| 0.1 | 3.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.3 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 6.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.8 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 7.8 | GO:0006935 | chemotaxis(GO:0006935) |
| 0.0 | 1.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.9 | 119.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 7.5 | 22.6 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 6.3 | 38.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 5.8 | 58.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 3.7 | 15.0 | GO:0000802 | transverse filament(GO:0000802) |
| 2.9 | 31.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.9 | 13.6 | GO:0070187 | telosome(GO:0070187) |
| 1.7 | 29.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.6 | 4.9 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 1.3 | 5.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 1.3 | 5.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.3 | 3.8 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 1.1 | 15.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.8 | 3.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.8 | 3.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.7 | 10.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.7 | 2.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.7 | 15.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.6 | 4.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.6 | 11.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 6.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 16.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.4 | 7.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 16.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 4.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 62.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.4 | 5.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 18.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 1.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 10.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 40.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 2.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 71.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 3.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 11.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 33.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 12.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 3.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 11.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 2.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 112.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 43.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 3.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 32.1 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 4.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 64.7 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 4.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 7.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.9 | 119.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 7.4 | 22.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 7.1 | 49.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 4.9 | 24.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 4.5 | 13.6 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 3.6 | 14.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 3.5 | 102.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 3.4 | 17.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 3.1 | 27.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 3.1 | 33.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 2.7 | 31.8 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 2.6 | 10.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 2.6 | 10.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.4 | 67.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 2.4 | 14.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 2.3 | 32.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 2.2 | 19.8 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 2.1 | 6.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.1 | 16.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.0 | 10.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.9 | 5.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 1.3 | 7.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.3 | 162.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 1.1 | 15.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.0 | 49.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 1.0 | 7.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.9 | 7.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.9 | 5.2 | GO:1902282 | phosphorelay sensor kinase activity(GO:0000155) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.8 | 7.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.7 | 10.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.7 | 52.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.7 | 12.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.6 | 4.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.6 | 3.0 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.6 | 9.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.5 | 4.8 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.5 | 30.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.5 | 3.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.4 | 2.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 13.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 5.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.4 | 1.5 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.4 | 5.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 72.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 12.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.3 | 2.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 9.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 5.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 2.4 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 3.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 13.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 28.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 36.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.2 | 17.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 6.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 22.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 3.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 2.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 17.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 3.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 4.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 1.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 10.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 12.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 6.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 41.2 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 10.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 35.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 5.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 11.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 5.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.5 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 14.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 22.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 3.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 30.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.1 | 37.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.8 | 32.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.6 | 27.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 16.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 40.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.4 | 10.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.4 | 97.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 13.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.4 | 10.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 4.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 18.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.3 | 7.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 5.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 19.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 6.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 3.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 5.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 55.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.7 | 55.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 1.5 | 27.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 1.3 | 37.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.1 | 44.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 1.0 | 24.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.8 | 27.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.8 | 33.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 6.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.6 | 18.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 10.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.5 | 4.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 4.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.5 | 10.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.5 | 22.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 16.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 7.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.4 | 7.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 19.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 2.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 37.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 5.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 15.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 9.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.2 | 25.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 27.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 5.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.5 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 2.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |