Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
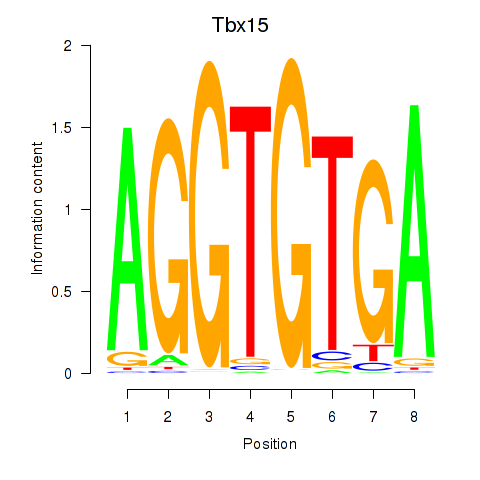
Results for Tbx15
Z-value: 0.56
Transcription factors associated with Tbx15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx15
|
ENSMUSG00000027868.5 | T-box 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx15 | mm10_v2_chr3_+_99253754_99253807 | 0.05 | 7.7e-01 | Click! |
Activity profile of Tbx15 motif
Sorted Z-values of Tbx15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_105933832 | 2.03 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr7_-_142679533 | 1.96 |
ENSMUST00000162317.1
ENSMUST00000125933.1 ENSMUST00000105931.1 ENSMUST00000105930.1 ENSMUST00000105933.1 ENSMUST00000105932.1 ENSMUST00000000220.2 |
Ins2
|
insulin II |
| chr17_-_31144271 | 1.38 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr6_-_116716888 | 0.84 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr4_+_148000722 | 0.72 |
ENSMUST00000103230.4
|
Nppa
|
natriuretic peptide type A |
| chr9_-_45204083 | 0.64 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr16_-_32797413 | 0.59 |
ENSMUST00000115116.1
ENSMUST00000041123.8 |
Muc20
|
mucin 20 |
| chr7_+_45705088 | 0.59 |
ENSMUST00000080885.3
|
Dbp
|
D site albumin promoter binding protein |
| chr8_+_45885479 | 0.57 |
ENSMUST00000034053.5
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr14_+_53683593 | 0.55 |
ENSMUST00000103663.4
|
Trav4-4-dv10
|
T cell receptor alpha variable 4-4-DV10 |
| chr6_-_113434757 | 0.55 |
ENSMUST00000113091.1
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr2_+_74681991 | 0.54 |
ENSMUST00000142312.1
|
Hoxd11
|
homeobox D11 |
| chr12_+_109459843 | 0.53 |
ENSMUST00000173812.1
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr3_-_65529355 | 0.52 |
ENSMUST00000099076.3
|
4931440P22Rik
|
RIKEN cDNA 4931440P22 gene |
| chr11_-_69900949 | 0.51 |
ENSMUST00000102580.3
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr7_+_28071230 | 0.51 |
ENSMUST00000138392.1
ENSMUST00000076648.7 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr2_-_13011747 | 0.50 |
ENSMUST00000061545.5
|
C1ql3
|
C1q-like 3 |
| chr6_+_78370877 | 0.50 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chrX_+_42151002 | 0.48 |
ENSMUST00000123245.1
|
Stag2
|
stromal antigen 2 |
| chr10_-_127341583 | 0.47 |
ENSMUST00000026474.3
|
Gli1
|
GLI-Kruppel family member GLI1 |
| chrX_+_7919816 | 0.47 |
ENSMUST00000041096.3
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr8_+_122422020 | 0.45 |
ENSMUST00000050963.3
|
Il17c
|
interleukin 17C |
| chr1_+_166130238 | 0.43 |
ENSMUST00000060833.7
ENSMUST00000166159.1 |
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr17_-_43502773 | 0.41 |
ENSMUST00000024707.8
ENSMUST00000117137.1 |
Mep1a
|
meprin 1 alpha |
| chr8_-_8639363 | 0.41 |
ENSMUST00000152698.1
|
Efnb2
|
ephrin B2 |
| chr7_+_30291659 | 0.39 |
ENSMUST00000014065.8
ENSMUST00000150892.1 ENSMUST00000126216.1 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr6_+_15196949 | 0.39 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr5_-_24351604 | 0.38 |
ENSMUST00000036092.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr16_-_43979050 | 0.37 |
ENSMUST00000165648.1
ENSMUST00000036321.7 |
Zdhhc23
|
zinc finger, DHHC domain containing 23 |
| chr13_+_99184733 | 0.37 |
ENSMUST00000056558.8
|
Zfp366
|
zinc finger protein 366 |
| chr4_+_84884418 | 0.37 |
ENSMUST00000169371.2
|
Cntln
|
centlein, centrosomal protein |
| chr19_-_42202150 | 0.37 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr10_-_88605017 | 0.36 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr8_+_84148252 | 0.36 |
ENSMUST00000093375.4
|
4930432K21Rik
|
RIKEN cDNA 4930432K21 gene |
| chr10_+_75566257 | 0.36 |
ENSMUST00000129232.1
ENSMUST00000143792.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr15_+_37233036 | 0.35 |
ENSMUST00000161405.1
ENSMUST00000022895.8 ENSMUST00000161532.1 |
Grhl2
|
grainyhead-like 2 (Drosophila) |
| chr1_+_166130467 | 0.34 |
ENSMUST00000166860.1
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr5_-_147307264 | 0.34 |
ENSMUST00000031650.3
|
Cdx2
|
caudal type homeobox 2 |
| chr11_-_120731944 | 0.34 |
ENSMUST00000154565.1
ENSMUST00000026148.2 |
Cbr2
|
carbonyl reductase 2 |
| chr7_-_126625676 | 0.34 |
ENSMUST00000032961.3
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr7_-_6730412 | 0.34 |
ENSMUST00000051209.4
|
Peg3
|
paternally expressed 3 |
| chr14_-_70524068 | 0.33 |
ENSMUST00000022692.3
|
Sftpc
|
surfactant associated protein C |
| chr14_+_65266701 | 0.32 |
ENSMUST00000169656.1
|
Fbxo16
|
F-box protein 16 |
| chr4_-_129121889 | 0.32 |
ENSMUST00000139450.1
ENSMUST00000125931.1 ENSMUST00000116444.2 |
Hpca
|
hippocalcin |
| chr17_+_29114142 | 0.32 |
ENSMUST00000141797.1
ENSMUST00000132262.1 ENSMUST00000141239.1 ENSMUST00000138816.1 |
Gm16194
|
predicted gene 16194 |
| chr19_+_53460610 | 0.31 |
ENSMUST00000180442.1
|
4833407H14Rik
|
RIKEN cDNA 4833407H14 gene |
| chr11_+_67171095 | 0.31 |
ENSMUST00000018641.7
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr9_-_61976563 | 0.31 |
ENSMUST00000113990.1
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr4_+_84884276 | 0.31 |
ENSMUST00000047023.6
|
Cntln
|
centlein, centrosomal protein |
| chr14_-_51057242 | 0.31 |
ENSMUST00000089798.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr5_+_3571664 | 0.31 |
ENSMUST00000008451.5
|
1700109H08Rik
|
RIKEN cDNA 1700109H08 gene |
| chr14_-_51988829 | 0.30 |
ENSMUST00000181008.1
|
Gm16617
|
predicted gene, 16617 |
| chr6_+_7555053 | 0.29 |
ENSMUST00000090679.2
ENSMUST00000184986.1 |
Tac1
|
tachykinin 1 |
| chr11_+_67171027 | 0.29 |
ENSMUST00000170159.1
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr19_-_10304867 | 0.29 |
ENSMUST00000039327.4
|
Dagla
|
diacylglycerol lipase, alpha |
| chr13_+_48662989 | 0.29 |
ENSMUST00000021813.4
|
Barx1
|
BarH-like homeobox 1 |
| chr11_-_70352029 | 0.28 |
ENSMUST00000019068.6
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr7_+_44496588 | 0.28 |
ENSMUST00000107927.3
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr8_+_84148025 | 0.28 |
ENSMUST00000143833.1
ENSMUST00000118856.1 |
4930432K21Rik
|
RIKEN cDNA 4930432K21 gene |
| chr1_-_75133866 | 0.28 |
ENSMUST00000027405.4
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr13_-_38151792 | 0.27 |
ENSMUST00000078232.1
|
Gm10129
|
predicted gene 10129 |
| chr14_+_45351473 | 0.27 |
ENSMUST00000111835.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chrX_+_109196750 | 0.27 |
ENSMUST00000139259.1
ENSMUST00000060013.3 |
Gm6377
|
predicted gene 6377 |
| chrX_-_162829379 | 0.27 |
ENSMUST00000041370.4
ENSMUST00000112316.2 ENSMUST00000112315.1 |
Txlng
|
taxilin gamma |
| chr3_+_95124476 | 0.27 |
ENSMUST00000131597.1
ENSMUST00000005769.6 ENSMUST00000107227.1 |
Tmod4
|
tropomodulin 4 |
| chr2_+_122637844 | 0.27 |
ENSMUST00000047498.8
|
AA467197
|
expressed sequence AA467197 |
| chr3_+_96181151 | 0.27 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr11_-_33578933 | 0.27 |
ENSMUST00000020366.1
ENSMUST00000135350.1 |
Gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr2_+_164562579 | 0.26 |
ENSMUST00000017867.3
ENSMUST00000109344.2 ENSMUST00000109345.2 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr15_-_79164477 | 0.26 |
ENSMUST00000040019.4
|
Sox10
|
SRY-box containing gene 10 |
| chr11_-_69900886 | 0.26 |
ENSMUST00000108621.2
ENSMUST00000100969.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr11_+_96323253 | 0.26 |
ENSMUST00000093944.3
|
Hoxb3
|
homeobox B3 |
| chr1_-_133025330 | 0.26 |
ENSMUST00000067429.3
ENSMUST00000067398.6 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr13_+_113209659 | 0.25 |
ENSMUST00000038144.8
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr4_-_141398204 | 0.25 |
ENSMUST00000105790.1
|
Clcnka
|
chloride channel Ka |
| chr11_+_3330781 | 0.25 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr5_-_24329556 | 0.25 |
ENSMUST00000115098.2
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr15_+_10952332 | 0.25 |
ENSMUST00000022853.8
ENSMUST00000110523.1 |
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr7_+_4119556 | 0.25 |
ENSMUST00000079415.5
|
Ttyh1
|
tweety homolog 1 (Drosophila) |
| chr4_-_57300751 | 0.24 |
ENSMUST00000151964.1
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr5_+_142702091 | 0.24 |
ENSMUST00000058418.7
|
Slc29a4
|
solute carrier family 29 (nucleoside transporters), member 4 |
| chr17_-_63863791 | 0.24 |
ENSMUST00000050753.3
|
A930002H24Rik
|
RIKEN cDNA A930002H24 gene |
| chr19_+_8617991 | 0.24 |
ENSMUST00000010250.2
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr5_-_8367982 | 0.24 |
ENSMUST00000088761.4
ENSMUST00000115386.1 ENSMUST00000050166.7 ENSMUST00000046838.7 ENSMUST00000115388.2 ENSMUST00000088744.5 ENSMUST00000115385.1 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr12_-_113422730 | 0.24 |
ENSMUST00000177715.1
ENSMUST00000103426.1 |
Ighm
|
immunoglobulin heavy constant mu |
| chr6_-_135118240 | 0.23 |
ENSMUST00000032327.7
ENSMUST00000111922.1 |
Gprc5d
|
G protein-coupled receptor, family C, group 5, member D |
| chr6_+_114131229 | 0.23 |
ENSMUST00000032451.7
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
| chr16_+_17276291 | 0.23 |
ENSMUST00000164950.1
ENSMUST00000159242.1 |
Tmem191c
|
transmembrane protein 191C |
| chrX_+_136666375 | 0.23 |
ENSMUST00000060904.4
ENSMUST00000113100.1 ENSMUST00000128040.1 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr6_-_113434529 | 0.23 |
ENSMUST00000133348.1
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr11_-_106301801 | 0.23 |
ENSMUST00000103071.3
|
Gh
|
growth hormone |
| chr11_-_94507337 | 0.23 |
ENSMUST00000040692.8
|
Mycbpap
|
MYCBP associated protein |
| chr16_+_17276337 | 0.23 |
ENSMUST00000159065.1
ENSMUST00000159494.1 ENSMUST00000159811.1 |
Tmem191c
|
transmembrane protein 191C |
| chr10_+_123264076 | 0.23 |
ENSMUST00000050756.7
|
Fam19a2
|
family with sequence similarity 19, member A2 |
| chr17_+_84511832 | 0.22 |
ENSMUST00000047206.5
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr7_-_30169701 | 0.22 |
ENSMUST00000062181.7
|
Zfp146
|
zinc finger protein 146 |
| chrX_+_139217166 | 0.22 |
ENSMUST00000166444.1
ENSMUST00000170671.1 ENSMUST00000113041.2 ENSMUST00000113042.2 |
Mum1l1
|
melanoma associated antigen (mutated) 1-like 1 |
| chrX_+_42149534 | 0.22 |
ENSMUST00000127618.1
|
Stag2
|
stromal antigen 2 |
| chr11_-_69900930 | 0.22 |
ENSMUST00000018714.6
ENSMUST00000128046.1 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr9_-_122862128 | 0.22 |
ENSMUST00000056467.7
|
Zfp445
|
zinc finger protein 445 |
| chr7_-_24316590 | 0.21 |
ENSMUST00000108436.1
ENSMUST00000032673.8 |
Zfp94
|
zinc finger protein 94 |
| chr18_-_47333311 | 0.21 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr7_-_78577771 | 0.21 |
ENSMUST00000039438.7
|
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr5_-_108749448 | 0.21 |
ENSMUST00000068946.7
|
Rnf212
|
ring finger protein 212 |
| chr11_+_29172890 | 0.21 |
ENSMUST00000102856.2
ENSMUST00000020755.5 |
Smek2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium) |
| chr3_-_89773221 | 0.21 |
ENSMUST00000038450.1
|
4632404H12Rik
|
RIKEN cDNA 4632404H12 gene |
| chr18_-_37644185 | 0.21 |
ENSMUST00000066272.4
|
Taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chrX_+_134295225 | 0.21 |
ENSMUST00000037687.7
|
Tmem35
|
transmembrane protein 35 |
| chr11_-_99993992 | 0.20 |
ENSMUST00000105049.1
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr4_-_41517326 | 0.20 |
ENSMUST00000030152.6
ENSMUST00000095126.4 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr8_-_40634750 | 0.20 |
ENSMUST00000173957.1
|
Mtmr7
|
myotubularin related protein 7 |
| chr14_+_52016849 | 0.20 |
ENSMUST00000100638.2
|
Tmem253
|
transmembrane protein 253 |
| chrX_-_7947553 | 0.20 |
ENSMUST00000133349.1
|
Hdac6
|
histone deacetylase 6 |
| chr6_+_86628174 | 0.19 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr11_+_43682038 | 0.19 |
ENSMUST00000094294.4
|
Pwwp2a
|
PWWP domain containing 2A |
| chr3_+_33799791 | 0.19 |
ENSMUST00000099153.3
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr6_+_115134899 | 0.19 |
ENSMUST00000009538.5
ENSMUST00000169345.1 |
Syn2
|
synapsin II |
| chrX_-_162964557 | 0.19 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr14_+_14012491 | 0.19 |
ENSMUST00000022257.2
|
Atxn7
|
ataxin 7 |
| chr8_-_4259257 | 0.19 |
ENSMUST00000053252.7
|
Ctxn1
|
cortexin 1 |
| chr5_+_114003678 | 0.19 |
ENSMUST00000112292.2
|
Dao
|
D-amino acid oxidase |
| chr9_-_49798905 | 0.18 |
ENSMUST00000114476.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr9_-_44721383 | 0.18 |
ENSMUST00000148929.1
ENSMUST00000123406.1 |
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr5_+_115908644 | 0.18 |
ENSMUST00000141101.1
|
Cit
|
citron |
| chr4_-_42773993 | 0.18 |
ENSMUST00000095114.4
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
| chr8_-_80739497 | 0.18 |
ENSMUST00000043359.8
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr7_+_4119525 | 0.18 |
ENSMUST00000119661.1
ENSMUST00000129423.1 |
Ttyh1
|
tweety homolog 1 (Drosophila) |
| chr10_+_86705811 | 0.18 |
ENSMUST00000061458.7
ENSMUST00000075632.6 |
BC030307
|
cDNA sequence BC030307 |
| chr1_+_93861344 | 0.18 |
ENSMUST00000094663.2
|
Gal3st2
|
galactose-3-O-sulfotransferase 2 |
| chr17_-_29237759 | 0.18 |
ENSMUST00000137727.1
ENSMUST00000024805.7 |
Cpne5
|
copine V |
| chr8_+_79028587 | 0.18 |
ENSMUST00000119254.1
|
Zfp827
|
zinc finger protein 827 |
| chr7_-_132813715 | 0.18 |
ENSMUST00000134946.1
|
Fam53b
|
family with sequence similarity 53, member B |
| chr5_-_116024475 | 0.17 |
ENSMUST00000111999.1
|
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr2_+_119174483 | 0.17 |
ENSMUST00000069711.2
|
Gm14137
|
predicted gene 14137 |
| chr11_+_83302817 | 0.17 |
ENSMUST00000142680.1
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr6_+_61180313 | 0.17 |
ENSMUST00000126214.1
|
Ccser1
|
coiled-coil serine rich 1 |
| chr6_-_145865302 | 0.17 |
ENSMUST00000111703.1
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr2_-_148046896 | 0.17 |
ENSMUST00000172928.1
ENSMUST00000047315.3 |
Foxa2
|
forkhead box A2 |
| chr2_-_91183017 | 0.17 |
ENSMUST00000066420.5
|
Madd
|
MAP-kinase activating death domain |
| chr17_+_35135463 | 0.17 |
ENSMUST00000173535.1
ENSMUST00000173952.1 |
Bag6
|
BCL2-associated athanogene 6 |
| chr11_+_96292453 | 0.17 |
ENSMUST00000173432.1
|
Hoxb6
|
homeobox B6 |
| chr13_+_81657732 | 0.17 |
ENSMUST00000049055.6
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr1_+_90953108 | 0.16 |
ENSMUST00000166281.1
|
Prlh
|
prolactin releasing hormone |
| chr3_+_145292472 | 0.16 |
ENSMUST00000029848.4
ENSMUST00000139001.1 |
Col24a1
|
collagen, type XXIV, alpha 1 |
| chr1_+_40580227 | 0.16 |
ENSMUST00000027233.7
|
Slc9a4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4 |
| chr17_-_71459300 | 0.16 |
ENSMUST00000183937.1
|
Gm4707
|
predicted gene 4707 |
| chr6_-_28261907 | 0.16 |
ENSMUST00000115320.1
ENSMUST00000123098.1 ENSMUST00000115321.2 ENSMUST00000155494.1 |
Zfp800
|
zinc finger protein 800 |
| chr15_-_88954400 | 0.16 |
ENSMUST00000109371.1
|
Ttll8
|
tubulin tyrosine ligase-like family, member 8 |
| chr2_+_144033059 | 0.16 |
ENSMUST00000037722.2
ENSMUST00000110032.1 |
Banf2
|
barrier to autointegration factor 2 |
| chr8_+_12915879 | 0.16 |
ENSMUST00000110876.2
ENSMUST00000110879.2 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr6_-_127769427 | 0.16 |
ENSMUST00000032500.8
|
Prmt8
|
protein arginine N-methyltransferase 8 |
| chr12_+_33314375 | 0.15 |
ENSMUST00000155386.1
|
Atxn7l1
|
ataxin 7-like 1 |
| chr3_+_98222148 | 0.15 |
ENSMUST00000029469.4
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr7_-_100658364 | 0.15 |
ENSMUST00000107043.1
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr9_+_71215779 | 0.15 |
ENSMUST00000034723.5
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr12_+_88953399 | 0.15 |
ENSMUST00000057634.7
|
Nrxn3
|
neurexin III |
| chr4_-_11386679 | 0.15 |
ENSMUST00000043781.7
ENSMUST00000108310.1 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr14_-_55116935 | 0.15 |
ENSMUST00000022819.5
|
Jph4
|
junctophilin 4 |
| chr3_-_89913144 | 0.15 |
ENSMUST00000029559.6
|
Il6ra
|
interleukin 6 receptor, alpha |
| chr2_-_132145057 | 0.15 |
ENSMUST00000028815.8
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr6_+_72304592 | 0.15 |
ENSMUST00000183018.1
|
Sftpb
|
surfactant associated protein B |
| chrX_-_7947763 | 0.15 |
ENSMUST00000154244.1
|
Hdac6
|
histone deacetylase 6 |
| chr17_-_6827990 | 0.14 |
ENSMUST00000181895.1
|
Gm2885
|
predicted gene 2885 |
| chr15_+_103503261 | 0.14 |
ENSMUST00000023132.3
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr1_-_105356658 | 0.14 |
ENSMUST00000058688.5
ENSMUST00000172299.1 |
Rnf152
|
ring finger protein 152 |
| chr11_+_43681998 | 0.14 |
ENSMUST00000061070.5
|
Pwwp2a
|
PWWP domain containing 2A |
| chr2_+_164823001 | 0.14 |
ENSMUST00000132282.1
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr5_-_31048014 | 0.14 |
ENSMUST00000137223.1
|
Slc5a6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6 |
| chr4_-_57300362 | 0.14 |
ENSMUST00000153926.1
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr14_+_53716680 | 0.13 |
ENSMUST00000103641.4
|
Trav7-6
|
T cell receptor alpha variable 7-6 |
| chr7_-_4604041 | 0.13 |
ENSMUST00000166650.1
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr11_+_104577281 | 0.13 |
ENSMUST00000106956.3
|
Myl4
|
myosin, light polypeptide 4 |
| chrX_-_150657392 | 0.13 |
ENSMUST00000151403.2
ENSMUST00000087253.4 ENSMUST00000112709.1 ENSMUST00000163969.1 ENSMUST00000087258.3 |
Tro
|
trophinin |
| chrX_+_166238923 | 0.13 |
ENSMUST00000060210.7
ENSMUST00000112233.1 |
Gpm6b
|
glycoprotein m6b |
| chr3_-_30140407 | 0.13 |
ENSMUST00000108271.3
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr4_-_11386757 | 0.13 |
ENSMUST00000108313.1
ENSMUST00000108311.2 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr11_+_104576965 | 0.13 |
ENSMUST00000106957.1
|
Myl4
|
myosin, light polypeptide 4 |
| chr9_+_46998931 | 0.13 |
ENSMUST00000178065.1
|
Gm4791
|
predicted gene 4791 |
| chr7_+_30169861 | 0.13 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr7_+_18987518 | 0.13 |
ENSMUST00000063563.7
|
Nanos2
|
nanos homolog 2 (Drosophila) |
| chr5_+_120649188 | 0.13 |
ENSMUST00000156722.1
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr12_-_111813834 | 0.12 |
ENSMUST00000021715.5
|
Xrcc3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr11_-_116654245 | 0.12 |
ENSMUST00000021166.5
|
Cygb
|
cytoglobin |
| chr16_+_93683184 | 0.12 |
ENSMUST00000039620.6
|
Cbr3
|
carbonyl reductase 3 |
| chr6_+_121343385 | 0.12 |
ENSMUST00000168295.1
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr16_+_87553313 | 0.12 |
ENSMUST00000026700.7
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr5_-_148399901 | 0.12 |
ENSMUST00000048116.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_-_116024452 | 0.12 |
ENSMUST00000031486.7
|
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr14_-_59142886 | 0.11 |
ENSMUST00000022548.3
ENSMUST00000162674.1 ENSMUST00000159858.1 ENSMUST00000162271.1 |
1700129C05Rik
|
RIKEN cDNA 1700129C05 gene |
| chr16_+_84835070 | 0.11 |
ENSMUST00000009120.7
|
Gabpa
|
GA repeat binding protein, alpha |
| chr8_-_68121527 | 0.11 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chr4_-_119658781 | 0.11 |
ENSMUST00000106309.2
ENSMUST00000044426.7 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr9_+_95954744 | 0.11 |
ENSMUST00000034981.7
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr11_-_57832679 | 0.11 |
ENSMUST00000160392.2
ENSMUST00000108845.2 |
Hand1
|
heart and neural crest derivatives expressed transcript 1 |
| chr4_+_84884366 | 0.11 |
ENSMUST00000102819.3
|
Cntln
|
centlein, centrosomal protein |
| chr14_+_53337122 | 0.11 |
ENSMUST00000179580.1
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr5_+_120513102 | 0.11 |
ENSMUST00000111889.1
|
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr17_-_81649607 | 0.11 |
ENSMUST00000163680.2
ENSMUST00000086538.3 ENSMUST00000163123.1 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr5_+_143622466 | 0.11 |
ENSMUST00000177196.1
|
Cyth3
|
cytohesin 3 |
| chr16_+_84834901 | 0.11 |
ENSMUST00000114184.1
|
Gabpa
|
GA repeat binding protein, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.2 | 1.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 0.7 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.2 | 0.6 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.4 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.5 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 0.3 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.1 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 0.2 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.3 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.1 | 0.3 | GO:0098917 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 0.3 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.1 | 0.2 | GO:0046436 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.2 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.5 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.4 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.2 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.2 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 | 0.2 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.0 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0006524 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.3 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.1 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.4 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.0 | GO:1904209 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.9 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 2.0 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.6 | GO:1902282 | phosphorelay sensor kinase activity(GO:0000155) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.3 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.2 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 0.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 0.3 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |