Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
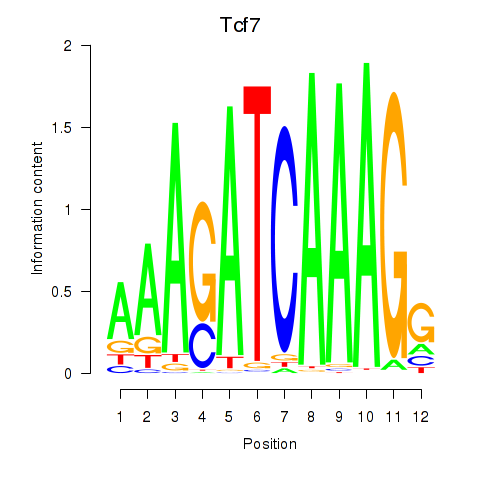
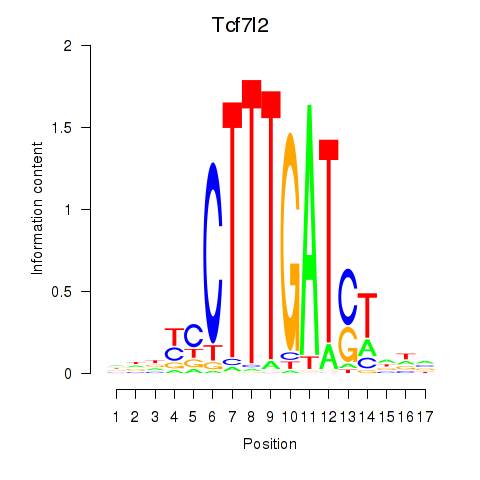
Results for Tcf7_Tcf7l2
Z-value: 1.24
Transcription factors associated with Tcf7_Tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7
|
ENSMUSG00000000782.9 | transcription factor 7, T cell specific |
|
Tcf7l2
|
ENSMUSG00000024985.12 | transcription factor 7 like 2, T cell specific, HMG box |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7 | mm10_v2_chr11_-_52282564_52282579 | 0.77 | 4.3e-08 | Click! |
| Tcf7l2 | mm10_v2_chr19_+_55894508_55894534 | 0.55 | 4.8e-04 | Click! |
Activity profile of Tcf7_Tcf7l2 motif
Sorted Z-values of Tcf7_Tcf7l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_145805862 | 9.49 |
ENSMUST00000067479.5
|
Cyp3a44
|
cytochrome P450, family 3, subfamily a, polypeptide 44 |
| chr14_-_56062307 | 8.01 |
ENSMUST00000043249.8
|
Mcpt4
|
mast cell protease 4 |
| chr5_-_145469723 | 6.58 |
ENSMUST00000031633.4
|
Cyp3a16
|
cytochrome P450, family 3, subfamily a, polypeptide 16 |
| chr6_-_40999479 | 5.33 |
ENSMUST00000166306.1
|
Gm2663
|
predicted gene 2663 |
| chr7_+_13733502 | 4.65 |
ENSMUST00000086148.6
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr3_-_107239707 | 4.49 |
ENSMUST00000049852.8
|
Prok1
|
prokineticin 1 |
| chr17_+_25366550 | 4.25 |
ENSMUST00000069616.7
|
Tpsb2
|
tryptase beta 2 |
| chr7_-_13989588 | 3.83 |
ENSMUST00000165167.1
ENSMUST00000108520.2 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr8_-_111691002 | 3.71 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr11_+_108920800 | 3.65 |
ENSMUST00000140821.1
|
Axin2
|
axin2 |
| chr5_-_145584723 | 3.51 |
ENSMUST00000075837.6
|
Cyp3a41b
|
cytochrome P450, family 3, subfamily a, polypeptide 41B |
| chr7_+_13623967 | 3.19 |
ENSMUST00000108525.2
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr18_+_21072329 | 2.94 |
ENSMUST00000082235.4
|
Mep1b
|
meprin 1 beta |
| chr7_-_142576492 | 2.94 |
ENSMUST00000140716.1
|
H19
|
H19 fetal liver mRNA |
| chr19_+_38132767 | 2.69 |
ENSMUST00000025956.5
ENSMUST00000112329.1 |
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr8_+_71597648 | 2.66 |
ENSMUST00000143662.1
|
Fam129c
|
family with sequence similarity 129, member C |
| chr6_-_69400097 | 2.50 |
ENSMUST00000177795.1
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr9_+_46240696 | 2.48 |
ENSMUST00000034585.6
|
Apoa4
|
apolipoprotein A-IV |
| chr7_-_142657466 | 2.43 |
ENSMUST00000097936.2
ENSMUST00000000033.5 |
Igf2
|
insulin-like growth factor 2 |
| chr6_+_129591782 | 2.33 |
ENSMUST00000112063.2
ENSMUST00000032268.7 ENSMUST00000119520.1 |
Klrd1
|
killer cell lectin-like receptor, subfamily D, member 1 |
| chr6_+_115422040 | 2.28 |
ENSMUST00000000450.3
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr9_-_96719549 | 2.24 |
ENSMUST00000128269.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr11_-_99322943 | 2.12 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr7_-_142656018 | 2.04 |
ENSMUST00000178921.1
|
Igf2
|
insulin-like growth factor 2 |
| chrX_+_6415736 | 2.00 |
ENSMUST00000143641.3
|
Shroom4
|
shroom family member 4 |
| chr7_+_45216671 | 1.99 |
ENSMUST00000134420.1
|
Tead2
|
TEA domain family member 2 |
| chr2_+_96318014 | 1.88 |
ENSMUST00000135431.1
ENSMUST00000162807.2 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr1_+_88166004 | 1.88 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr14_-_70323783 | 1.83 |
ENSMUST00000151011.1
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_-_150255591 | 1.83 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
| chr10_-_128960965 | 1.75 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr6_+_78405148 | 1.75 |
ENSMUST00000023906.2
|
Reg2
|
regenerating islet-derived 2 |
| chr7_-_31076656 | 1.72 |
ENSMUST00000167369.1
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr16_-_45492962 | 1.71 |
ENSMUST00000114585.2
|
Gm609
|
predicted gene 609 |
| chr5_-_44102032 | 1.67 |
ENSMUST00000171543.1
|
Prom1
|
prominin 1 |
| chr6_-_69284319 | 1.66 |
ENSMUST00000103349.1
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr8_+_21734490 | 1.65 |
ENSMUST00000080533.5
|
Defa24
|
defensin, alpha, 24 |
| chr19_+_39992424 | 1.64 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr5_-_62765618 | 1.64 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_+_40964760 | 1.59 |
ENSMUST00000076638.5
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr13_+_65512678 | 1.56 |
ENSMUST00000081471.2
|
Gm10139
|
predicted gene 10139 |
| chr10_-_127370535 | 1.54 |
ENSMUST00000026472.8
|
Inhbc
|
inhibin beta-C |
| chr19_+_18749983 | 1.53 |
ENSMUST00000040489.7
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr11_+_108921648 | 1.51 |
ENSMUST00000144511.1
|
Axin2
|
axin2 |
| chr17_+_8236031 | 1.48 |
ENSMUST00000164411.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr14_+_63436394 | 1.48 |
ENSMUST00000121288.1
|
Fam167a
|
family with sequence similarity 167, member A |
| chr13_+_4434306 | 1.48 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr13_-_66851513 | 1.47 |
ENSMUST00000169322.1
|
Gm17404
|
predicted gene, 17404 |
| chr10_-_13388830 | 1.44 |
ENSMUST00000079698.5
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr15_-_54278420 | 1.41 |
ENSMUST00000079772.3
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr3_+_57425314 | 1.41 |
ENSMUST00000029377.7
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr4_+_44943727 | 1.37 |
ENSMUST00000154177.1
|
Gm12678
|
predicted gene 12678 |
| chr15_+_9436028 | 1.35 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr2_-_62483637 | 1.35 |
ENSMUST00000136686.1
ENSMUST00000102733.3 |
Gcg
|
glucagon |
| chr11_+_67277124 | 1.34 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr3_-_96814518 | 1.34 |
ENSMUST00000047702.7
|
Cd160
|
CD160 antigen |
| chr17_-_78684262 | 1.34 |
ENSMUST00000145480.1
|
Strn
|
striatin, calmodulin binding protein |
| chr2_-_164221605 | 1.32 |
ENSMUST00000018355.4
ENSMUST00000109376.2 |
Wfdc15b
|
WAP four-disulfide core domain 15B |
| chr8_+_109990430 | 1.31 |
ENSMUST00000001720.7
ENSMUST00000143741.1 |
Tat
|
tyrosine aminotransferase |
| chr12_-_41485751 | 1.29 |
ENSMUST00000043884.4
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr1_+_131638306 | 1.28 |
ENSMUST00000073350.6
|
Ctse
|
cathepsin E |
| chr19_-_39886730 | 1.28 |
ENSMUST00000168838.1
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr6_-_69243445 | 1.26 |
ENSMUST00000101325.3
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr9_-_96719404 | 1.21 |
ENSMUST00000140121.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr10_+_127849917 | 1.20 |
ENSMUST00000077530.2
|
Rdh19
|
retinol dehydrogenase 19 |
| chr2_-_150179679 | 1.17 |
ENSMUST00000099418.2
|
Gm10770
|
predicted gene 10770 |
| chr13_+_94083490 | 1.17 |
ENSMUST00000156071.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr6_+_129045893 | 1.17 |
ENSMUST00000032257.7
|
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F |
| chr4_-_134254076 | 1.15 |
ENSMUST00000060050.5
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr11_+_108920342 | 1.13 |
ENSMUST00000052915.7
|
Axin2
|
axin2 |
| chr4_+_141420757 | 1.12 |
ENSMUST00000102486.4
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr10_-_13388753 | 1.11 |
ENSMUST00000105546.1
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr9_-_123862023 | 1.08 |
ENSMUST00000182350.1
ENSMUST00000078755.2 |
Xcr1
|
chemokine (C motif) receptor 1 |
| chrX_-_150812932 | 1.06 |
ENSMUST00000131241.1
ENSMUST00000147152.1 ENSMUST00000143843.1 |
Maged2
|
melanoma antigen, family D, 2 |
| chr19_+_8591254 | 1.02 |
ENSMUST00000010251.3
ENSMUST00000170817.1 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr3_+_55242526 | 0.99 |
ENSMUST00000054237.7
ENSMUST00000167204.1 |
Dclk1
|
doublecortin-like kinase 1 |
| chr8_-_95294074 | 0.98 |
ENSMUST00000184103.1
|
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr7_+_128062657 | 0.97 |
ENSMUST00000120355.1
ENSMUST00000106240.2 ENSMUST00000098015.3 |
Itgam
|
integrin alpha M |
| chr14_+_45219993 | 0.96 |
ENSMUST00000146150.1
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr1_-_139560158 | 0.96 |
ENSMUST00000160423.1
ENSMUST00000023965.5 |
Cfhr1
|
complement factor H-related 1 |
| chr9_+_77921908 | 0.95 |
ENSMUST00000133757.1
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr13_+_54701457 | 0.95 |
ENSMUST00000037145.7
|
Cdhr2
|
cadherin-related family member 2 |
| chr9_+_75232013 | 0.94 |
ENSMUST00000036555.6
|
Myo5c
|
myosin VC |
| chr5_-_145879857 | 0.94 |
ENSMUST00000035918.7
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr5_-_87569023 | 0.92 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr7_-_48558066 | 0.89 |
ENSMUST00000052730.1
|
Mrgprb2
|
MAS-related GPR, member B2 |
| chr13_-_56548534 | 0.88 |
ENSMUST00000062806.4
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_-_134225088 | 0.86 |
ENSMUST00000067680.4
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr10_-_115587739 | 0.86 |
ENSMUST00000020350.8
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr9_+_51280295 | 0.85 |
ENSMUST00000050829.1
|
2010007H06Rik
|
RIKEN cDNA 2010007H06 gene |
| chr11_-_60036917 | 0.85 |
ENSMUST00000102692.3
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr12_+_24831583 | 0.83 |
ENSMUST00000110942.3
ENSMUST00000078902.6 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr14_-_45219364 | 0.83 |
ENSMUST00000022377.4
ENSMUST00000143609.1 ENSMUST00000139526.1 |
Txndc16
|
thioredoxin domain containing 16 |
| chr6_-_87335758 | 0.81 |
ENSMUST00000042025.9
|
Antxr1
|
anthrax toxin receptor 1 |
| chr10_-_22820126 | 0.80 |
ENSMUST00000049930.7
|
Tcf21
|
transcription factor 21 |
| chr4_+_95967322 | 0.80 |
ENSMUST00000107083.1
|
Hook1
|
hook homolog 1 (Drosophila) |
| chr5_+_99979061 | 0.79 |
ENSMUST00000046721.1
|
4930524J08Rik
|
RIKEN cDNA 4930524J08 gene |
| chr17_-_47016956 | 0.78 |
ENSMUST00000165525.1
|
Gm16494
|
predicted gene 16494 |
| chr14_+_101840501 | 0.78 |
ENSMUST00000159026.1
|
Lmo7
|
LIM domain only 7 |
| chr16_-_44139630 | 0.75 |
ENSMUST00000137557.1
ENSMUST00000147025.1 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chrX_-_150812715 | 0.73 |
ENSMUST00000112697.3
|
Maged2
|
melanoma antigen, family D, 2 |
| chr15_+_102296256 | 0.72 |
ENSMUST00000064924.4
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae) |
| chr13_-_66852017 | 0.72 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chr14_+_51114455 | 0.72 |
ENSMUST00000100645.3
|
Eddm3b
|
epididymal protein 3B |
| chr10_+_21978643 | 0.72 |
ENSMUST00000142174.1
ENSMUST00000164659.1 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_-_47528862 | 0.72 |
ENSMUST00000172559.1
|
Mrgpra2b
|
MAS-related GPR, member A2B |
| chr1_+_88095054 | 0.72 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr13_+_47122719 | 0.71 |
ENSMUST00000068891.4
|
Rnf144b
|
ring finger protein 144B |
| chr1_+_88087802 | 0.71 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr13_-_41828418 | 0.70 |
ENSMUST00000137905.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr3_-_59130610 | 0.69 |
ENSMUST00000065220.6
ENSMUST00000091112.4 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr3_+_55461758 | 0.68 |
ENSMUST00000070418.4
|
Dclk1
|
doublecortin-like kinase 1 |
| chr7_-_45211877 | 0.68 |
ENSMUST00000033057.7
|
Dkkl1
|
dickkopf-like 1 |
| chr19_+_58670358 | 0.68 |
ENSMUST00000057270.7
|
Pnlip
|
pancreatic lipase |
| chr7_+_38183787 | 0.67 |
ENSMUST00000067854.8
ENSMUST00000177983.1 |
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr8_-_86580664 | 0.67 |
ENSMUST00000131423.1
ENSMUST00000152438.1 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr4_-_19922599 | 0.67 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr4_+_108479081 | 0.67 |
ENSMUST00000155068.1
|
Zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr8_+_21665797 | 0.67 |
ENSMUST00000075268.4
|
Gm15315
|
predicted gene 15315 |
| chr7_+_132859225 | 0.67 |
ENSMUST00000084497.5
ENSMUST00000181577.1 ENSMUST00000106161.1 |
Fam175b
|
family with sequence similarity 175, member B |
| chr10_+_115569986 | 0.66 |
ENSMUST00000173620.1
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr6_+_117168535 | 0.64 |
ENSMUST00000112866.1
ENSMUST00000112871.1 ENSMUST00000073043.4 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr1_-_139781236 | 0.64 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr2_-_91931675 | 0.63 |
ENSMUST00000111309.1
|
Mdk
|
midkine |
| chr1_+_172126750 | 0.63 |
ENSMUST00000075895.2
ENSMUST00000111252.3 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr1_+_88227005 | 0.62 |
ENSMUST00000061013.6
ENSMUST00000113130.1 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr11_+_108381164 | 0.62 |
ENSMUST00000146050.1
ENSMUST00000152958.1 |
Apoh
|
apolipoprotein H |
| chr4_-_43429071 | 0.62 |
ENSMUST00000107929.3
ENSMUST00000107928.2 |
Fam166b
|
family with sequence similarity 166, member B |
| chr13_+_23870284 | 0.62 |
ENSMUST00000006785.7
|
Slc17a1
|
solute carrier family 17 (sodium phosphate), member 1 |
| chr2_-_91931696 | 0.62 |
ENSMUST00000090602.5
|
Mdk
|
midkine |
| chr4_-_142239356 | 0.61 |
ENSMUST00000036476.3
|
Kazn
|
kazrin, periplakin interacting protein |
| chr1_+_88070765 | 0.61 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_+_157560078 | 0.61 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr13_-_62858364 | 0.60 |
ENSMUST00000021907.7
|
Fbp2
|
fructose bisphosphatase 2 |
| chr1_-_133906973 | 0.60 |
ENSMUST00000126123.1
|
Optc
|
opticin |
| chr12_+_38783455 | 0.60 |
ENSMUST00000161980.1
ENSMUST00000160701.1 |
Etv1
|
ets variant gene 1 |
| chr8_+_21565368 | 0.59 |
ENSMUST00000084040.2
|
Gm14850
|
predicted gene 14850 |
| chr15_-_78495059 | 0.59 |
ENSMUST00000089398.1
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr12_+_4133394 | 0.58 |
ENSMUST00000152065.1
ENSMUST00000127756.1 |
Adcy3
|
adenylate cyclase 3 |
| chr1_-_75133866 | 0.58 |
ENSMUST00000027405.4
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chrY_+_90755657 | 0.58 |
ENSMUST00000167967.2
|
Gm21857
|
predicted gene, 21857 |
| chr12_+_4133047 | 0.58 |
ENSMUST00000124505.1
|
Adcy3
|
adenylate cyclase 3 |
| chr13_-_22219820 | 0.58 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr15_-_66286224 | 0.57 |
ENSMUST00000070256.7
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr15_+_10981747 | 0.56 |
ENSMUST00000070877.5
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr10_+_127000709 | 0.56 |
ENSMUST00000026500.5
ENSMUST00000142698.1 |
Avil
|
advillin |
| chr8_-_8639363 | 0.56 |
ENSMUST00000152698.1
|
Efnb2
|
ephrin B2 |
| chr6_+_54267131 | 0.56 |
ENSMUST00000114402.2
|
Chn2
|
chimerin (chimaerin) 2 |
| chr13_-_107890059 | 0.56 |
ENSMUST00000105097.2
|
Zswim6
|
zinc finger SWIM-type containing 6 |
| chr1_+_157506728 | 0.55 |
ENSMUST00000086130.2
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr2_+_70474923 | 0.54 |
ENSMUST00000100043.2
|
Sp5
|
trans-acting transcription factor 5 |
| chr1_+_157506777 | 0.54 |
ENSMUST00000027881.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr6_+_117559694 | 0.54 |
ENSMUST00000162741.1
ENSMUST00000008011.7 |
Gm9946
|
predicted gene 9946 |
| chr1_-_168432270 | 0.54 |
ENSMUST00000072863.4
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr1_+_88055467 | 0.53 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr3_-_15838643 | 0.53 |
ENSMUST00000148194.1
|
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr5_-_105239533 | 0.52 |
ENSMUST00000065588.6
|
Gbp10
|
guanylate-binding protein 10 |
| chr16_+_29210108 | 0.52 |
ENSMUST00000162747.1
|
Hrasls
|
HRAS-like suppressor |
| chr5_+_17574268 | 0.52 |
ENSMUST00000030568.7
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr5_+_86071734 | 0.52 |
ENSMUST00000031171.7
|
Stap1
|
signal transducing adaptor family member 1 |
| chr10_+_38965515 | 0.51 |
ENSMUST00000019992.5
|
Lama4
|
laminin, alpha 4 |
| chr14_+_54431597 | 0.51 |
ENSMUST00000089688.4
|
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr3_+_118562129 | 0.50 |
ENSMUST00000039177.7
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr4_+_121039385 | 0.50 |
ENSMUST00000030372.5
|
Col9a2
|
collagen, type IX, alpha 2 |
| chr4_-_43429117 | 0.50 |
ENSMUST00000171134.2
ENSMUST00000052829.3 |
Fam166b
|
family with sequence similarity 166, member B |
| chr11_+_96929367 | 0.49 |
ENSMUST00000062172.5
|
Prr15l
|
proline rich 15-like |
| chr4_-_16013796 | 0.48 |
ENSMUST00000149891.1
|
Osgin2
|
oxidative stress induced growth inhibitor family member 2 |
| chr18_+_37400845 | 0.48 |
ENSMUST00000057228.1
|
Pcdhb9
|
protocadherin beta 9 |
| chr6_+_72629284 | 0.47 |
ENSMUST00000156497.1
|
Gm15401
|
predicted gene 15401 |
| chr3_-_116712696 | 0.47 |
ENSMUST00000169530.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr10_+_69787431 | 0.46 |
ENSMUST00000183240.1
|
Ank3
|
ankyrin 3, epithelial |
| chr6_+_86628174 | 0.46 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr4_+_45848816 | 0.46 |
ENSMUST00000107782.1
ENSMUST00000030011.5 |
1300002K09Rik
|
RIKEN cDNA 1300002K09 gene |
| chr14_+_56017964 | 0.46 |
ENSMUST00000022836.4
|
Mcpt1
|
mast cell protease 1 |
| chr18_+_44334062 | 0.46 |
ENSMUST00000025349.5
ENSMUST00000115498.1 |
Myot
|
myotilin |
| chr8_+_21297439 | 0.45 |
ENSMUST00000098892.4
|
Defa5
|
defensin, alpha, 5 |
| chr4_-_106727930 | 0.45 |
ENSMUST00000106770.1
ENSMUST00000145044.1 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chrX_+_169879596 | 0.45 |
ENSMUST00000112105.1
ENSMUST00000078947.5 |
Mid1
|
midline 1 |
| chr13_+_42301270 | 0.44 |
ENSMUST00000021796.7
|
Edn1
|
endothelin 1 |
| chr5_-_108795352 | 0.44 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr7_+_128062635 | 0.44 |
ENSMUST00000064821.7
ENSMUST00000106242.3 |
Itgam
|
integrin alpha M |
| chr8_-_107065632 | 0.44 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr12_-_87299705 | 0.43 |
ENSMUST00000125733.1
|
Ism2
|
isthmin 2 homolog (zebrafish) |
| chr4_+_32238713 | 0.43 |
ENSMUST00000108180.2
|
Bach2
|
BTB and CNC homology 2 |
| chr1_+_88055377 | 0.42 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr6_-_71322200 | 0.42 |
ENSMUST00000173297.1
ENSMUST00000114188.2 |
Smyd1
|
SET and MYND domain containing 1 |
| chr7_-_3845050 | 0.41 |
ENSMUST00000108615.3
ENSMUST00000119469.1 |
Pira2
|
paired-Ig-like receptor A2 |
| chr14_+_53665912 | 0.40 |
ENSMUST00000181768.1
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chr9_-_71896047 | 0.40 |
ENSMUST00000184448.1
|
Tcf12
|
transcription factor 12 |
| chr9_-_98033181 | 0.40 |
ENSMUST00000035027.6
|
Clstn2
|
calsyntenin 2 |
| chr7_+_140247316 | 0.39 |
ENSMUST00000164583.1
ENSMUST00000093984.2 |
5830411N06Rik
|
RIKEN cDNA 5830411N06 gene |
| chr1_+_139454747 | 0.39 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr17_-_40794063 | 0.39 |
ENSMUST00000131699.1
ENSMUST00000024724.7 ENSMUST00000144243.1 |
Crisp2
|
cysteine-rich secretory protein 2 |
| chr2_-_35061431 | 0.38 |
ENSMUST00000028233.3
|
Hc
|
hemolytic complement |
| chr1_+_171840557 | 0.38 |
ENSMUST00000135386.1
|
Cd84
|
CD84 antigen |
| chr11_-_110251736 | 0.38 |
ENSMUST00000044003.7
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr4_+_136206365 | 0.37 |
ENSMUST00000047526.7
|
Asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr13_+_45389734 | 0.37 |
ENSMUST00000038275.9
|
Mylip
|
myosin regulatory light chain interacting protein |
| chr10_+_89744988 | 0.37 |
ENSMUST00000020112.5
|
Uhrf1bp1l
|
UHRF1 (ICBP90) binding protein 1-like |
| chr19_-_40073731 | 0.37 |
ENSMUST00000048959.3
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7_Tcf7l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.8 | 2.5 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 0.8 | 2.3 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.6 | 8.0 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.6 | 4.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 1.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 1.5 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 4.9 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 1.5 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.3 | 1.0 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 | 0.8 | GO:0060435 | bronchiole development(GO:0060435) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 1.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.7 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 0.7 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.2 | 2.9 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 1.8 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.8 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 2.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 1.2 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 1.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 0.9 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 0.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.3 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 0.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.9 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 0.2 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.2 | 0.5 | GO:0046104 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) thymidine metabolic process(GO:0046104) |
| 0.1 | 0.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.4 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.8 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 2.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.4 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 1.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.6 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 3.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.3 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.5 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.6 | GO:0034197 | blood coagulation, intrinsic pathway(GO:0007597) acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 1.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.6 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 1.3 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.7 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.2 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 2.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.6 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.3 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.6 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 1.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.2 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.1 | 2.0 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.1 | 2.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.7 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.7 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0050883 | regulation of sulfur amino acid metabolic process(GO:0031335) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 1.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.2 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 1.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.3 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 5.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 2.0 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.6 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 4.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 1.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.4 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 1.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 2.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0051596 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.5 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 1.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.0 | 0.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 2.7 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 0.9 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 1.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 3.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 1.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 18.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 2.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 4.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 7.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 27.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 20.5 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 1.0 | 7.0 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.8 | 2.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.5 | 1.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 1.3 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 6.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 2.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 1.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 0.6 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 1.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 2.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 0.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 3.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 5.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 4.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 23.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 3.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.0 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.3 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.1 | 1.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.5 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 3.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.2 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.8 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 8.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.1 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 1.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 3.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 1.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 3.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 3.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 3.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 2.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 4.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |