Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
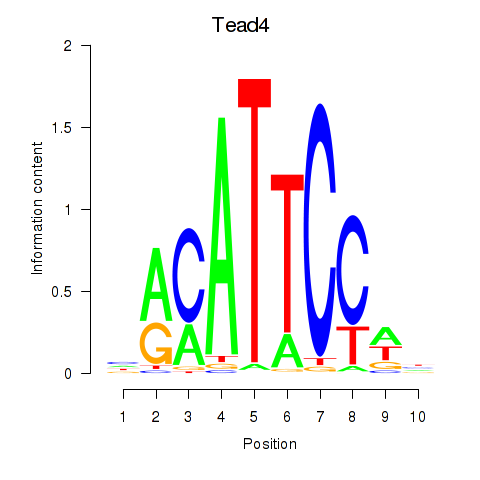
Results for Tead3_Tead4
Z-value: 1.65
Transcription factors associated with Tead3_Tead4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tead3
|
ENSMUSG00000002249.11 | TEA domain family member 3 |
|
Tead4
|
ENSMUSG00000030353.9 | TEA domain family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead4 | mm10_v2_chr6_-_128300738_128300823 | 0.79 | 1.1e-08 | Click! |
| Tead3 | mm10_v2_chr17_-_28350747_28350820 | 0.52 | 1.0e-03 | Click! |
Activity profile of Tead3_Tead4 motif
Sorted Z-values of Tead3_Tead4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_43558386 | 15.14 |
ENSMUST00000130353.1
|
Tln1
|
talin 1 |
| chr16_+_38362205 | 12.58 |
ENSMUST00000023494.6
|
Popdc2
|
popeye domain containing 2 |
| chr16_+_38362234 | 9.95 |
ENSMUST00000114739.1
|
Popdc2
|
popeye domain containing 2 |
| chr8_-_85365317 | 9.15 |
ENSMUST00000034133.7
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_85365341 | 8.96 |
ENSMUST00000121972.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr17_-_35702297 | 8.02 |
ENSMUST00000135078.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr3_-_90695706 | 7.63 |
ENSMUST00000069960.5
ENSMUST00000117167.1 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr7_-_24760311 | 7.34 |
ENSMUST00000063956.5
|
Cd177
|
CD177 antigen |
| chr19_-_34255325 | 7.27 |
ENSMUST00000039631.8
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr6_-_67535783 | 7.22 |
ENSMUST00000058178.4
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr7_-_132813799 | 7.10 |
ENSMUST00000097998.2
|
Fam53b
|
family with sequence similarity 53, member B |
| chr1_+_43730593 | 6.82 |
ENSMUST00000027217.8
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene |
| chr8_-_123894736 | 6.63 |
ENSMUST00000034453.4
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chrX_+_136224035 | 6.61 |
ENSMUST00000113116.2
|
Tceal7
|
transcription elongation factor A (SII)-like 7 |
| chr7_-_34133215 | 6.34 |
ENSMUST00000038537.8
|
Wtip
|
WT1-interacting protein |
| chr12_-_76709997 | 5.73 |
ENSMUST00000166101.1
|
Sptb
|
spectrin beta, erythrocytic |
| chr11_-_116077927 | 5.64 |
ENSMUST00000156545.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr5_+_76840597 | 5.46 |
ENSMUST00000120639.2
ENSMUST00000163347.1 ENSMUST00000121851.1 |
C530008M17Rik
|
RIKEN cDNA C530008M17 gene |
| chr17_+_71019548 | 5.44 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr18_+_50030977 | 5.22 |
ENSMUST00000145726.1
ENSMUST00000128377.1 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr4_-_118457450 | 5.20 |
ENSMUST00000106375.1
ENSMUST00000006556.3 ENSMUST00000168404.1 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr6_-_115758974 | 5.12 |
ENSMUST00000072933.6
|
Tmem40
|
transmembrane protein 40 |
| chr17_+_71019503 | 5.11 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr19_+_38132767 | 5.10 |
ENSMUST00000025956.5
ENSMUST00000112329.1 |
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr18_+_61953048 | 4.97 |
ENSMUST00000051720.5
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr4_-_118457509 | 4.87 |
ENSMUST00000102671.3
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr11_-_116077954 | 4.72 |
ENSMUST00000106451.1
ENSMUST00000075036.2 |
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr14_-_54994541 | 4.66 |
ENSMUST00000153783.1
ENSMUST00000168485.1 ENSMUST00000102803.3 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr11_-_116077562 | 4.65 |
ENSMUST00000174822.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr4_-_88033328 | 4.62 |
ENSMUST00000078090.5
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr7_-_44524642 | 4.53 |
ENSMUST00000165208.2
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr17_-_35701937 | 4.52 |
ENSMUST00000155628.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr11_+_104577281 | 4.39 |
ENSMUST00000106956.3
|
Myl4
|
myosin, light polypeptide 4 |
| chr7_-_132813715 | 4.38 |
ENSMUST00000134946.1
|
Fam53b
|
family with sequence similarity 53, member B |
| chr17_-_35702040 | 4.11 |
ENSMUST00000166980.2
ENSMUST00000145900.1 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chrX_-_142306170 | 4.10 |
ENSMUST00000134825.2
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr14_-_54966570 | 3.94 |
ENSMUST00000124930.1
ENSMUST00000134256.1 ENSMUST00000081857.6 ENSMUST00000145322.1 |
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr18_+_60911757 | 3.87 |
ENSMUST00000040359.5
|
Arsi
|
arylsulfatase i |
| chr18_+_23415400 | 3.83 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr1_+_135836380 | 3.74 |
ENSMUST00000178204.1
|
Tnnt2
|
troponin T2, cardiac |
| chr11_+_62077018 | 3.73 |
ENSMUST00000092415.5
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr2_+_91035613 | 3.73 |
ENSMUST00000111445.3
ENSMUST00000111446.3 ENSMUST00000050323.5 |
Rapsn
|
receptor-associated protein of the synapse |
| chr11_+_104576965 | 3.69 |
ENSMUST00000106957.1
|
Myl4
|
myosin, light polypeptide 4 |
| chr1_+_189728264 | 3.62 |
ENSMUST00000097442.2
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr11_+_44519405 | 3.58 |
ENSMUST00000101327.2
|
Rnf145
|
ring finger protein 145 |
| chr8_-_122551316 | 3.55 |
ENSMUST00000067252.7
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr12_+_85746539 | 3.44 |
ENSMUST00000040461.3
|
Mfsd7c
|
major facilitator superfamily domain containing 7C |
| chr9_-_18512885 | 3.42 |
ENSMUST00000034653.6
|
Muc16
|
mucin 16 |
| chr7_-_38107490 | 3.40 |
ENSMUST00000108023.3
|
Ccne1
|
cyclin E1 |
| chr15_-_58324161 | 3.37 |
ENSMUST00000022985.1
|
Klhl38
|
kelch-like 38 |
| chr4_+_49059256 | 3.36 |
ENSMUST00000076670.2
|
E130309F12Rik
|
RIKEN cDNA E130309F12 gene |
| chr3_+_153844209 | 3.31 |
ENSMUST00000044089.3
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr18_-_60501983 | 3.31 |
ENSMUST00000042710.6
|
Smim3
|
small integral membrane protein 3 |
| chr18_+_50053282 | 3.27 |
ENSMUST00000148159.2
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr4_-_134018829 | 3.26 |
ENSMUST00000051674.2
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr4_-_4138432 | 3.15 |
ENSMUST00000070375.7
|
Penk
|
preproenkephalin |
| chr7_+_26061495 | 3.05 |
ENSMUST00000005669.7
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chrX_-_49788204 | 3.05 |
ENSMUST00000114893.1
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr9_+_69454066 | 2.99 |
ENSMUST00000134907.1
|
Anxa2
|
annexin A2 |
| chr17_+_47593516 | 2.96 |
ENSMUST00000182874.1
|
Ccnd3
|
cyclin D3 |
| chr11_+_98412461 | 2.92 |
ENSMUST00000058295.5
|
Erbb2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) |
| chr2_-_156839790 | 2.92 |
ENSMUST00000134838.1
ENSMUST00000137463.1 ENSMUST00000149275.2 |
Gm14230
|
predicted gene 14230 |
| chr4_+_154237525 | 2.91 |
ENSMUST00000152159.1
|
Megf6
|
multiple EGF-like-domains 6 |
| chr14_-_69284982 | 2.84 |
ENSMUST00000183882.1
ENSMUST00000037064.4 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr8_-_104624266 | 2.80 |
ENSMUST00000163783.2
|
Cdh16
|
cadherin 16 |
| chr13_-_117025505 | 2.80 |
ENSMUST00000022239.6
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_-_136835426 | 2.80 |
ENSMUST00000068293.3
ENSMUST00000111894.1 |
Smco3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr15_+_6386598 | 2.80 |
ENSMUST00000080880.5
ENSMUST00000110664.2 ENSMUST00000110663.2 ENSMUST00000161812.1 ENSMUST00000160134.1 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr1_-_66945019 | 2.77 |
ENSMUST00000027151.5
|
Myl1
|
myosin, light polypeptide 1 |
| chr8_+_57511833 | 2.75 |
ENSMUST00000067925.6
|
Hmgb2
|
high mobility group box 2 |
| chr4_+_134315112 | 2.74 |
ENSMUST00000105875.1
ENSMUST00000030638.6 |
Trim63
|
tripartite motif-containing 63 |
| chr11_-_100970887 | 2.73 |
ENSMUST00000060792.5
|
Ptrf
|
polymerase I and transcript release factor |
| chr14_-_69503220 | 2.65 |
ENSMUST00000180059.2
|
Gm21464
|
predicted gene, 21464 |
| chr1_-_66945361 | 2.61 |
ENSMUST00000160100.1
|
Myl1
|
myosin, light polypeptide 1 |
| chr14_-_69503316 | 2.49 |
ENSMUST00000179116.2
|
Gm21464
|
predicted gene, 21464 |
| chr2_-_92370999 | 2.46 |
ENSMUST00000176810.1
ENSMUST00000090582.4 |
Gyltl1b
|
glycosyltransferase-like 1B |
| chr6_-_128300738 | 2.44 |
ENSMUST00000143004.1
ENSMUST00000006311.6 ENSMUST00000112157.2 ENSMUST00000133118.1 |
Tead4
|
TEA domain family member 4 |
| chr9_-_95815389 | 2.44 |
ENSMUST00000119760.1
|
Pls1
|
plastin 1 (I-isoform) |
| chr6_+_29433248 | 2.42 |
ENSMUST00000101617.2
ENSMUST00000065090.5 |
Flnc
|
filamin C, gamma |
| chr17_+_47593444 | 2.40 |
ENSMUST00000182209.1
|
Ccnd3
|
cyclin D3 |
| chr16_-_42340595 | 2.40 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr2_-_92371039 | 2.39 |
ENSMUST00000068586.6
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr5_+_150259922 | 2.38 |
ENSMUST00000087204.5
|
Fry
|
furry homolog (Drosophila) |
| chr5_+_31116722 | 2.35 |
ENSMUST00000114637.1
|
Trim54
|
tripartite motif-containing 54 |
| chr5_+_31116702 | 2.35 |
ENSMUST00000013771.8
|
Trim54
|
tripartite motif-containing 54 |
| chr4_-_126163470 | 2.34 |
ENSMUST00000097891.3
|
Sh3d21
|
SH3 domain containing 21 |
| chr9_-_96437434 | 2.18 |
ENSMUST00000070500.2
|
BC043934
|
cDNA sequence BC043934 |
| chr4_+_120588728 | 2.18 |
ENSMUST00000171482.1
|
Gm8439
|
predicted gene 8439 |
| chr7_+_127211608 | 2.16 |
ENSMUST00000032910.6
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr15_+_54410755 | 2.10 |
ENSMUST00000036737.3
|
Colec10
|
collectin sub-family member 10 |
| chr17_-_71002488 | 2.10 |
ENSMUST00000148960.1
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_-_42276688 | 2.05 |
ENSMUST00000175881.1
ENSMUST00000056974.3 |
Foxo3
|
forkhead box O3 |
| chr16_+_26463135 | 2.04 |
ENSMUST00000161053.1
ENSMUST00000115302.1 |
Cldn16
|
claudin 16 |
| chrX_+_56454871 | 2.03 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr2_-_92370968 | 2.03 |
ENSMUST00000176774.1
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chrX_+_139217166 | 2.02 |
ENSMUST00000166444.1
ENSMUST00000170671.1 ENSMUST00000113041.2 ENSMUST00000113042.2 |
Mum1l1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr10_-_62814539 | 2.02 |
ENSMUST00000173087.1
ENSMUST00000174121.1 |
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr4_+_135120640 | 2.02 |
ENSMUST00000056977.7
|
Runx3
|
runt related transcription factor 3 |
| chrX_-_74246364 | 1.97 |
ENSMUST00000130007.1
|
Flna
|
filamin, alpha |
| chr17_+_8236031 | 1.95 |
ENSMUST00000164411.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr6_+_29433131 | 1.94 |
ENSMUST00000090474.4
|
Flnc
|
filamin C, gamma |
| chr16_+_30008657 | 1.93 |
ENSMUST00000181485.1
|
4632428C04Rik
|
RIKEN cDNA 4632428C04 gene |
| chr16_+_87553313 | 1.93 |
ENSMUST00000026700.7
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr11_-_5803733 | 1.88 |
ENSMUST00000020768.3
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr8_-_48555846 | 1.85 |
ENSMUST00000110345.1
ENSMUST00000110343.1 |
Tenm3
|
teneurin transmembrane protein 3 |
| chrX_-_74246534 | 1.83 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr3_+_19644452 | 1.82 |
ENSMUST00000029139.7
|
Trim55
|
tripartite motif-containing 55 |
| chr6_+_29694204 | 1.80 |
ENSMUST00000046750.7
ENSMUST00000115250.3 |
Tspan33
|
tetraspanin 33 |
| chr7_+_29303938 | 1.77 |
ENSMUST00000108231.1
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr6_+_87428986 | 1.74 |
ENSMUST00000032125.5
|
Bmp10
|
bone morphogenetic protein 10 |
| chr1_-_149922339 | 1.74 |
ENSMUST00000111926.2
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr2_-_58160495 | 1.73 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr3_-_116807733 | 1.70 |
ENSMUST00000159670.1
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr7_-_132317198 | 1.69 |
ENSMUST00000080215.5
|
Chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr1_-_126830632 | 1.68 |
ENSMUST00000112583.1
ENSMUST00000094609.3 |
Nckap5
|
NCK-associated protein 5 |
| chr13_+_55369732 | 1.66 |
ENSMUST00000063771.7
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr2_-_73486456 | 1.66 |
ENSMUST00000141264.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_51289106 | 1.63 |
ENSMUST00000051572.6
|
Sdpr
|
serum deprivation response |
| chr14_+_70890099 | 1.60 |
ENSMUST00000022699.8
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2 |
| chr5_-_69542622 | 1.58 |
ENSMUST00000031045.6
|
Yipf7
|
Yip1 domain family, member 7 |
| chr8_-_12573311 | 1.57 |
ENSMUST00000180858.1
|
D630011A20Rik
|
RIKEN cDNA D630011A20 gene |
| chr6_+_122513676 | 1.57 |
ENSMUST00000142896.1
ENSMUST00000121656.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr7_-_28372597 | 1.55 |
ENSMUST00000144700.1
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr4_-_11965699 | 1.55 |
ENSMUST00000108301.1
ENSMUST00000095144.3 ENSMUST00000108302.1 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chrX_+_96456362 | 1.54 |
ENSMUST00000079322.5
ENSMUST00000113838.1 |
Heph
|
hephaestin |
| chr19_+_33822908 | 1.54 |
ENSMUST00000042061.6
|
Gm5519
|
predicted pseudogene 5519 |
| chr11_+_100415697 | 1.52 |
ENSMUST00000001595.3
|
Fkbp10
|
FK506 binding protein 10 |
| chr9_+_69453620 | 1.51 |
ENSMUST00000034756.8
ENSMUST00000123470.1 |
Anxa2
|
annexin A2 |
| chr13_+_109926832 | 1.50 |
ENSMUST00000117420.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr4_-_132398199 | 1.49 |
ENSMUST00000136711.1
ENSMUST00000084249.4 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr11_+_100415722 | 1.48 |
ENSMUST00000107400.2
|
Fkbp10
|
FK506 binding protein 10 |
| chr13_-_113046357 | 1.47 |
ENSMUST00000022282.3
|
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr5_+_66968559 | 1.45 |
ENSMUST00000127184.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr6_+_122513643 | 1.42 |
ENSMUST00000118626.1
|
Mfap5
|
microfibrillar associated protein 5 |
| chr17_-_34615965 | 1.42 |
ENSMUST00000097345.3
ENSMUST00000015611.7 |
Egfl8
|
EGF-like domain 8 |
| chr15_-_79254737 | 1.40 |
ENSMUST00000039752.3
|
Slc16a8
|
solute carrier family 16 (monocarboxylic acid transporters), member 8 |
| chr2_+_156840077 | 1.39 |
ENSMUST00000081335.6
ENSMUST00000073352.3 |
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr8_+_121950492 | 1.38 |
ENSMUST00000093078.6
ENSMUST00000170857.1 ENSMUST00000026354.8 ENSMUST00000174753.1 ENSMUST00000172511.1 |
Banp
|
BTG3 associated nuclear protein |
| chr2_-_73485733 | 1.38 |
ENSMUST00000102680.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr6_-_28831747 | 1.37 |
ENSMUST00000062304.5
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr13_-_62858364 | 1.37 |
ENSMUST00000021907.7
|
Fbp2
|
fructose bisphosphatase 2 |
| chrX_-_12128386 | 1.37 |
ENSMUST00000145872.1
|
Bcor
|
BCL6 interacting corepressor |
| chr19_-_32210969 | 1.36 |
ENSMUST00000151289.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr2_-_148443543 | 1.36 |
ENSMUST00000099269.3
|
Cd93
|
CD93 antigen |
| chr16_+_11984581 | 1.35 |
ENSMUST00000170672.2
ENSMUST00000023138.7 |
Shisa9
|
shisa homolog 9 (Xenopus laevis) |
| chr15_-_42676967 | 1.34 |
ENSMUST00000022921.5
|
Angpt1
|
angiopoietin 1 |
| chr3_+_88364548 | 1.34 |
ENSMUST00000147948.1
ENSMUST00000147991.1 |
Paqr6
|
progestin and adipoQ receptor family member VI |
| chr1_+_156558759 | 1.33 |
ENSMUST00000173929.1
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr2_-_120404058 | 1.32 |
ENSMUST00000090042.5
ENSMUST00000110729.1 ENSMUST00000090046.5 |
Tmem87a
|
transmembrane protein 87A |
| chr1_+_171155512 | 1.31 |
ENSMUST00000111334.1
|
Mpz
|
myelin protein zero |
| chr17_+_34608997 | 1.29 |
ENSMUST00000114140.3
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr4_+_133176336 | 1.28 |
ENSMUST00000105912.1
|
Wasf2
|
WAS protein family, member 2 |
| chr1_-_136234113 | 1.27 |
ENSMUST00000120339.1
ENSMUST00000048668.8 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr18_+_82554463 | 1.26 |
ENSMUST00000062446.7
ENSMUST00000102812.4 ENSMUST00000075372.5 ENSMUST00000080658.4 ENSMUST00000152071.1 ENSMUST00000114674.3 ENSMUST00000142850.1 ENSMUST00000133193.1 ENSMUST00000123251.1 ENSMUST00000153478.1 ENSMUST00000132369.1 |
Mbp
|
myelin basic protein |
| chr2_+_121295437 | 1.26 |
ENSMUST00000110639.1
|
Map1a
|
microtubule-associated protein 1 A |
| chr6_+_41684414 | 1.25 |
ENSMUST00000031900.5
|
1700034O15Rik
|
RIKEN cDNA 1700034O15 gene |
| chr11_-_60913775 | 1.25 |
ENSMUST00000019075.3
|
Gm16515
|
predicted gene, Gm16515 |
| chr3_-_144819494 | 1.23 |
ENSMUST00000029929.7
|
Clca2
|
chloride channel calcium activated 2 |
| chr9_-_48340876 | 1.23 |
ENSMUST00000034527.7
|
Nxpe2
|
neurexophilin and PC-esterase domain family, member 2 |
| chrX_-_51681856 | 1.23 |
ENSMUST00000114871.1
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr11_+_101984270 | 1.22 |
ENSMUST00000176722.1
ENSMUST00000175972.1 |
1700006E09Rik
|
RIKEN cDNA 1700006E09 gene |
| chr1_-_153186447 | 1.21 |
ENSMUST00000027753.6
|
Lamc2
|
laminin, gamma 2 |
| chr6_+_17307040 | 1.21 |
ENSMUST00000123439.1
|
Cav1
|
caveolin 1, caveolae protein |
| chr7_-_28372494 | 1.21 |
ENSMUST00000119990.1
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr8_+_110266960 | 1.20 |
ENSMUST00000043141.6
|
Hydin
|
HYDIN, axonemal central pair apparatus protein |
| chr17_-_47834682 | 1.20 |
ENSMUST00000066368.6
|
Mdfi
|
MyoD family inhibitor |
| chrX_-_51681703 | 1.20 |
ENSMUST00000088172.5
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr11_+_87109221 | 1.19 |
ENSMUST00000020794.5
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_+_88532314 | 1.17 |
ENSMUST00000172699.1
|
Mex3a
|
mex3 homolog A (C. elegans) |
| chr12_-_55014329 | 1.17 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr17_-_35697971 | 1.17 |
ENSMUST00000146472.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr1_+_12692430 | 1.16 |
ENSMUST00000180062.1
ENSMUST00000177608.1 |
Sulf1
|
sulfatase 1 |
| chr11_-_67771115 | 1.16 |
ENSMUST00000051765.8
|
Glp2r
|
glucagon-like peptide 2 receptor |
| chr18_-_47333311 | 1.15 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_+_24595623 | 1.14 |
ENSMUST00000176228.1
ENSMUST00000129142.1 |
Ctgf
|
connective tissue growth factor |
| chr17_-_71002017 | 1.13 |
ENSMUST00000128179.1
ENSMUST00000150456.1 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr16_-_36784924 | 1.13 |
ENSMUST00000168279.1
ENSMUST00000164579.1 ENSMUST00000023616.2 |
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chrX_+_72987339 | 1.12 |
ENSMUST00000164800.1
ENSMUST00000114546.2 |
Zfp185
|
zinc finger protein 185 |
| chr11_+_96323253 | 1.12 |
ENSMUST00000093944.3
|
Hoxb3
|
homeobox B3 |
| chr15_-_36598019 | 1.12 |
ENSMUST00000155116.1
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr8_-_87804411 | 1.10 |
ENSMUST00000165770.2
|
Zfp423
|
zinc finger protein 423 |
| chr6_+_122513583 | 1.09 |
ENSMUST00000032210.7
ENSMUST00000148517.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr9_+_75071579 | 1.09 |
ENSMUST00000136731.1
|
Myo5a
|
myosin VA |
| chrX_+_6415736 | 1.08 |
ENSMUST00000143641.3
|
Shroom4
|
shroom family member 4 |
| chr16_-_95586585 | 1.08 |
ENSMUST00000077773.6
|
Erg
|
avian erythroblastosis virus E-26 (v-ets) oncogene related |
| chr3_+_37639985 | 1.08 |
ENSMUST00000108107.1
|
Spry1
|
sprouty homolog 1 (Drosophila) |
| chr2_+_30595037 | 1.07 |
ENSMUST00000102853.3
|
Cstad
|
CSA-conditional, T cell activation-dependent protein |
| chr11_+_69125896 | 1.07 |
ENSMUST00000021268.2
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr7_-_14562171 | 1.06 |
ENSMUST00000181796.1
|
Vmn1r90
|
vomeronasal 1 receptor 90 |
| chr15_+_98632220 | 1.06 |
ENSMUST00000109150.1
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr16_-_38800193 | 1.05 |
ENSMUST00000057767.4
|
Upk1b
|
uroplakin 1B |
| chr11_+_115974709 | 1.05 |
ENSMUST00000021107.7
ENSMUST00000106461.1 ENSMUST00000169928.1 |
Itgb4
|
integrin beta 4 |
| chr4_-_83052229 | 1.05 |
ENSMUST00000107230.1
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr13_-_77131276 | 1.05 |
ENSMUST00000159300.1
|
Ankrd32
|
ankyrin repeat domain 32 |
| chr4_-_154636831 | 1.04 |
ENSMUST00000030902.6
ENSMUST00000105637.1 ENSMUST00000070313.7 ENSMUST00000105636.1 ENSMUST00000105638.2 ENSMUST00000097759.2 ENSMUST00000124771.1 |
Prdm16
|
PR domain containing 16 |
| chr16_+_77014069 | 1.02 |
ENSMUST00000023580.6
|
Usp25
|
ubiquitin specific peptidase 25 |
| chr6_-_142507805 | 1.02 |
ENSMUST00000134191.1
ENSMUST00000032373.5 |
Ldhb
|
lactate dehydrogenase B |
| chr4_-_83052147 | 1.02 |
ENSMUST00000170248.2
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr19_-_57197496 | 1.02 |
ENSMUST00000111544.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr10_-_13324250 | 1.01 |
ENSMUST00000105543.1
|
Phactr2
|
phosphatase and actin regulator 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead3_Tead4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 22.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 3.4 | 10.1 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 3.0 | 18.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 2.5 | 7.6 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 1.9 | 15.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.7 | 15.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 1.5 | 17.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.3 | 3.8 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.2 | 3.7 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.1 | 5.6 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 1.1 | 6.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 1.0 | 15.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.9 | 4.7 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.9 | 2.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.9 | 3.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.8 | 7.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.8 | 3.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.8 | 6.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.7 | 4.5 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.7 | 2.9 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.7 | 4.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.6 | 2.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 9.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.6 | 1.7 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.5 | 3.2 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.5 | 2.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.5 | 2.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.4 | 1.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.4 | 3.4 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.4 | 5.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.4 | 1.3 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.4 | 2.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 1.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 2.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.4 | 3.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 1.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 1.5 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 1.9 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.4 | 3.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 2.5 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.3 | 1.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 2.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.3 | 2.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.3 | 1.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.3 | 1.6 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.3 | 2.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 5.0 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 1.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 1.7 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 0.9 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 1.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 2.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 0.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.9 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.2 | 1.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 1.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 4.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.2 | 1.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.4 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 1.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 0.8 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 | 1.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 0.7 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) pancreas morphogenesis(GO:0061113) |
| 0.2 | 2.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 0.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 0.5 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.2 | 1.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 0.5 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 | 0.7 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.2 | 1.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 7.8 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.2 | 0.8 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 1.7 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 1.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 0.5 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.2 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 5.8 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 4.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 8.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 5.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 9.8 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.1 | 1.0 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 1.6 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 | 2.0 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 0.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 2.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 1.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.8 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 1.3 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 3.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.8 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 2.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.4 | GO:0035522 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.0 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 | 1.9 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 3.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.8 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 3.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 1.0 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.1 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 2.0 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 0.5 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 4.9 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 1.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 1.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 3.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 1.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.4 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.9 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 1.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.3 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.7 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 3.9 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 2.1 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 1.4 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 1.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 1.4 | 15.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.0 | 7.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.9 | 22.9 | GO:0032982 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.8 | 5.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 3.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.8 | 3.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.7 | 2.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.6 | 3.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 4.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.4 | 1.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 1.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 1.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.3 | 6.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 2.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 1.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 0.8 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 14.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 4.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 2.9 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 2.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.6 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 6.9 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.2 | 0.5 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.2 | 1.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 12.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 4.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 9.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 3.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 2.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 22.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 4.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.8 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 20.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 14.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 3.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 5.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 5.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.1 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 6.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 2.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.0 | 17.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.3 | 10.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.3 | 15.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.0 | 4.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.0 | 2.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.9 | 13.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.9 | 4.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.8 | 3.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.7 | 2.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.6 | 1.9 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.6 | 2.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.5 | 1.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.5 | 3.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.5 | 21.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 4.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.4 | 2.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 1.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 1.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.4 | 1.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.4 | 1.9 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.4 | 3.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 3.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 2.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 5.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.3 | 5.1 | GO:0030553 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 2.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 1.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.3 | 1.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 10.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 1.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 3.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 4.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.8 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 3.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.7 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 2.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 0.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 2.7 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 8.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 9.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 0.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 6.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.9 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 2.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.5 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 3.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.7 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.7 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 1.9 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 2.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 8.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 2.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 13.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.7 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 1.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0052744 | phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 3.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 3.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 5.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 3.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.8 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 10.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 4.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 5.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 6.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 8.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 5.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 6.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 9.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 5.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 2.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 2.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.5 | 24.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 3.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 2.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 4.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 2.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 10.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.2 | 3.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 3.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 5.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 5.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 3.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 6.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 3.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.8 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |