Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
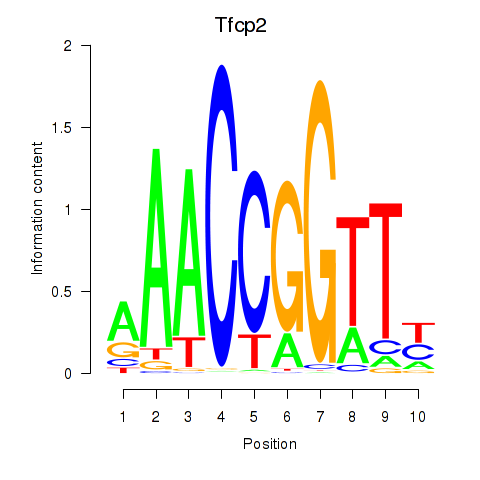
Results for Tfcp2
Z-value: 1.74
Transcription factors associated with Tfcp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2
|
ENSMUSG00000009733.8 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2 | mm10_v2_chr15_-_100551959_100552010 | 0.32 | 5.5e-02 | Click! |
Activity profile of Tfcp2 motif
Sorted Z-values of Tfcp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_75571522 | 16.60 |
ENSMUST00000143226.1
ENSMUST00000124259.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_+_57511833 | 16.03 |
ENSMUST00000067925.6
|
Hmgb2
|
high mobility group box 2 |
| chr19_-_34166037 | 13.74 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr8_+_94172618 | 13.02 |
ENSMUST00000034214.6
|
Mt2
|
metallothionein 2 |
| chr10_+_43579161 | 12.61 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr17_-_48451510 | 12.51 |
ENSMUST00000024794.5
|
Tspo2
|
translocator protein 2 |
| chr7_+_96211656 | 12.31 |
ENSMUST00000107165.1
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr11_+_116532441 | 11.34 |
ENSMUST00000106386.1
ENSMUST00000145737.1 ENSMUST00000155102.1 ENSMUST00000063446.6 |
Sphk1
|
sphingosine kinase 1 |
| chr5_+_106964319 | 10.85 |
ENSMUST00000031221.5
ENSMUST00000117196.2 ENSMUST00000076467.6 |
Cdc7
|
cell division cycle 7 (S. cerevisiae) |
| chr11_-_99438143 | 10.64 |
ENSMUST00000017743.2
|
Krt20
|
keratin 20 |
| chr3_-_90695706 | 10.36 |
ENSMUST00000069960.5
ENSMUST00000117167.1 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr11_+_69045640 | 9.80 |
ENSMUST00000108666.1
ENSMUST00000021277.5 |
Aurkb
|
aurora kinase B |
| chr15_+_89322969 | 9.46 |
ENSMUST00000066991.5
|
Adm2
|
adrenomedullin 2 |
| chrX_+_73639414 | 9.26 |
ENSMUST00000019701.8
|
Dusp9
|
dual specificity phosphatase 9 |
| chr17_+_28207778 | 9.15 |
ENSMUST00000002327.5
|
Def6
|
differentially expressed in FDCP 6 |
| chr11_+_11487671 | 8.52 |
ENSMUST00000020410.4
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chrX_-_102250940 | 8.40 |
ENSMUST00000134887.1
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr8_+_3665747 | 8.39 |
ENSMUST00000014118.2
|
1810033B17Rik
|
RIKEN cDNA 1810033B17 gene |
| chr15_-_103537928 | 8.12 |
ENSMUST00000023133.6
|
Ppp1r1a
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr6_+_134929089 | 7.78 |
ENSMUST00000183867.1
ENSMUST00000184991.1 ENSMUST00000183905.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr9_-_67832325 | 7.66 |
ENSMUST00000054500.5
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr6_+_134929118 | 7.64 |
ENSMUST00000185152.1
ENSMUST00000184504.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr6_+_5390387 | 7.29 |
ENSMUST00000183358.1
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr13_-_100786402 | 6.80 |
ENSMUST00000174038.1
ENSMUST00000091295.7 ENSMUST00000072119.8 |
Ccnb1
|
cyclin B1 |
| chr4_-_154025657 | 6.50 |
ENSMUST00000146426.1
|
Smim1
|
small integral membrane protein 1 |
| chr1_+_107511489 | 6.34 |
ENSMUST00000064916.2
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr5_+_33658550 | 6.33 |
ENSMUST00000152847.1
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr4_-_154025867 | 6.24 |
ENSMUST00000130175.1
ENSMUST00000182151.1 |
Smim1
|
small integral membrane protein 1 |
| chr1_+_107511416 | 6.21 |
ENSMUST00000009356.4
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr12_+_24708984 | 6.04 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr6_+_127887582 | 5.91 |
ENSMUST00000032501.4
|
Tspan11
|
tetraspanin 11 |
| chr4_-_154025926 | 5.82 |
ENSMUST00000132541.1
ENSMUST00000143047.1 |
Smim1
|
small integral membrane protein 1 |
| chr11_-_95914884 | 5.74 |
ENSMUST00000038343.6
|
B4galnt2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
| chr14_-_37110087 | 5.63 |
ENSMUST00000179488.1
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chrX_-_102250775 | 5.55 |
ENSMUST00000130589.1
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr4_-_49593875 | 5.55 |
ENSMUST00000151542.1
|
Tmem246
|
transmembrane protein 246 |
| chr5_+_33658567 | 5.27 |
ENSMUST00000114426.3
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr11_+_96929260 | 5.27 |
ENSMUST00000054311.5
ENSMUST00000107636.3 |
Prr15l
|
proline rich 15-like |
| chr17_-_25727364 | 5.06 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr11_+_23256001 | 4.86 |
ENSMUST00000020538.6
ENSMUST00000109551.1 ENSMUST00000102870.1 ENSMUST00000102869.1 |
Xpo1
|
exportin 1, CRM1 homolog (yeast) |
| chr13_+_91461050 | 4.82 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr2_+_85136355 | 4.82 |
ENSMUST00000057019.7
|
Aplnr
|
apelin receptor |
| chr1_+_130731963 | 4.79 |
ENSMUST00000039323.6
|
AA986860
|
expressed sequence AA986860 |
| chr19_-_40588374 | 4.78 |
ENSMUST00000175932.1
ENSMUST00000176955.1 ENSMUST00000149476.2 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr4_-_154025616 | 4.77 |
ENSMUST00000182191.1
ENSMUST00000146543.2 |
Smim1
|
small integral membrane protein 1 |
| chr7_-_4546567 | 4.69 |
ENSMUST00000065957.5
|
Syt5
|
synaptotagmin V |
| chr19_-_40588453 | 4.66 |
ENSMUST00000025979.6
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr15_-_74734313 | 4.60 |
ENSMUST00000023260.3
|
Lypd2
|
Ly6/Plaur domain containing 2 |
| chrX_-_164076100 | 4.56 |
ENSMUST00000037928.2
ENSMUST00000071667.2 |
Siah1b
|
seven in absentia 1B |
| chr19_-_40588338 | 4.40 |
ENSMUST00000176939.1
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr4_-_133967893 | 4.33 |
ENSMUST00000100472.3
ENSMUST00000136327.1 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_87156127 | 4.13 |
ENSMUST00000160810.1
|
Ecel1
|
endothelin converting enzyme-like 1 |
| chr11_+_69914179 | 4.12 |
ENSMUST00000057884.5
|
Gps2
|
G protein pathway suppressor 2 |
| chr9_+_124102110 | 4.09 |
ENSMUST00000168841.1
ENSMUST00000055918.6 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr18_+_11633276 | 4.05 |
ENSMUST00000115861.2
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr7_-_45333754 | 3.90 |
ENSMUST00000042194.8
|
Trpm4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr3_-_55055038 | 3.81 |
ENSMUST00000029368.2
|
Ccna1
|
cyclin A1 |
| chr11_+_3895223 | 3.79 |
ENSMUST00000055931.4
ENSMUST00000109996.1 |
Dusp18
|
dual specificity phosphatase 18 |
| chr8_+_69822429 | 3.77 |
ENSMUST00000164890.1
ENSMUST00000034325.4 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr14_-_73325773 | 3.72 |
ENSMUST00000022701.6
|
Rb1
|
retinoblastoma 1 |
| chr2_-_168010618 | 3.71 |
ENSMUST00000099073.2
|
Fam65c
|
family with sequence similarity 65, member C |
| chr14_-_76556662 | 3.62 |
ENSMUST00000064517.7
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr1_-_52190901 | 3.60 |
ENSMUST00000156887.1
ENSMUST00000129107.1 |
Gls
|
glutaminase |
| chr2_+_26628441 | 3.54 |
ENSMUST00000074240.3
|
Fam69b
|
family with sequence similarity 69, member B |
| chr15_-_73184840 | 3.44 |
ENSMUST00000044113.10
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr9_+_50856924 | 3.42 |
ENSMUST00000174628.1
ENSMUST00000034560.7 ENSMUST00000114437.2 ENSMUST00000175645.1 ENSMUST00000176349.1 ENSMUST00000176798.1 ENSMUST00000175640.1 |
Ppp2r1b
|
protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform |
| chrX_-_37085402 | 3.37 |
ENSMUST00000115231.3
|
Rpl39
|
ribosomal protein L39 |
| chr18_+_60925612 | 3.33 |
ENSMUST00000102888.3
ENSMUST00000025519.4 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr14_+_75136475 | 3.26 |
ENSMUST00000122840.1
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr4_-_116167591 | 3.23 |
ENSMUST00000030465.3
ENSMUST00000143426.1 |
Tspan1
|
tetraspanin 1 |
| chr4_-_133967953 | 3.21 |
ENSMUST00000102553.4
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_50376982 | 3.15 |
ENSMUST00000109142.1
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr3_+_122729158 | 3.12 |
ENSMUST00000066728.5
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chrX_-_75130914 | 3.10 |
ENSMUST00000114091.1
|
Mpp1
|
membrane protein, palmitoylated |
| chr9_+_124101944 | 3.09 |
ENSMUST00000171719.1
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr19_-_38043559 | 3.06 |
ENSMUST00000041475.8
ENSMUST00000172095.2 |
Myof
|
myoferlin |
| chrX_-_75130996 | 2.97 |
ENSMUST00000033775.2
|
Mpp1
|
membrane protein, palmitoylated |
| chr15_-_79164477 | 2.94 |
ENSMUST00000040019.4
|
Sox10
|
SRY-box containing gene 10 |
| chr17_-_24527925 | 2.93 |
ENSMUST00000176652.1
|
Traf7
|
TNF receptor-associated factor 7 |
| chrX_+_6779306 | 2.90 |
ENSMUST00000067410.7
|
Dgkk
|
diacylglycerol kinase kappa |
| chr16_-_32810477 | 2.83 |
ENSMUST00000179384.2
|
Gm933
|
predicted gene 933 |
| chr17_-_24527830 | 2.81 |
ENSMUST00000176353.1
ENSMUST00000176237.1 |
Traf7
|
TNF receptor-associated factor 7 |
| chr1_-_149961230 | 2.79 |
ENSMUST00000070200.8
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr19_-_60861390 | 2.69 |
ENSMUST00000135808.1
|
Sfxn4
|
sideroflexin 4 |
| chr2_-_51934943 | 2.69 |
ENSMUST00000102767.1
ENSMUST00000102768.1 |
Rbm43
|
RNA binding motif protein 43 |
| chr3_-_102964124 | 2.57 |
ENSMUST00000058899.8
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr8_+_120488416 | 2.55 |
ENSMUST00000034279.9
|
Gse1
|
genetic suppressor element 1 |
| chr1_+_58210397 | 2.51 |
ENSMUST00000040442.5
|
Aox4
|
aldehyde oxidase 4 |
| chr10_+_80019621 | 2.50 |
ENSMUST00000043311.6
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr2_+_84798828 | 2.47 |
ENSMUST00000102642.2
ENSMUST00000150325.1 |
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr2_-_180334665 | 2.45 |
ENSMUST00000015771.2
|
Gata5
|
GATA binding protein 5 |
| chr5_-_129787175 | 2.42 |
ENSMUST00000031399.6
|
Psph
|
phosphoserine phosphatase |
| chr8_-_110846770 | 2.40 |
ENSMUST00000042012.5
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chrX_+_56779437 | 2.40 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_+_114786045 | 2.40 |
ENSMUST00000137519.1
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr11_+_48800357 | 2.38 |
ENSMUST00000020640.7
|
Gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 like 1 |
| chr8_+_121730563 | 2.31 |
ENSMUST00000026357.5
|
Jph3
|
junctophilin 3 |
| chr6_+_122990367 | 2.29 |
ENSMUST00000079379.2
|
Clec4a4
|
C-type lectin domain family 4, member a4 |
| chr13_-_76018524 | 2.29 |
ENSMUST00000050997.1
ENSMUST00000179078.1 ENSMUST00000167271.1 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr10_+_127048235 | 2.26 |
ENSMUST00000165764.1
|
Cyp27b1
|
cytochrome P450, family 27, subfamily b, polypeptide 1 |
| chr17_+_35059035 | 2.21 |
ENSMUST00000007255.6
ENSMUST00000174493.1 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr15_-_100424208 | 2.20 |
ENSMUST00000154331.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr3_+_95622236 | 2.19 |
ENSMUST00000074353.4
|
Rps10-ps1
|
ribosomal protein S10, pseudogene 1 |
| chr19_-_29367294 | 2.16 |
ENSMUST00000138051.1
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr14_+_75136326 | 2.14 |
ENSMUST00000145303.1
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr19_-_36919606 | 2.11 |
ENSMUST00000057337.7
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr1_-_133690100 | 2.10 |
ENSMUST00000169295.1
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr16_-_3907651 | 2.09 |
ENSMUST00000177221.1
ENSMUST00000177323.1 |
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr7_-_35396708 | 2.08 |
ENSMUST00000154597.1
ENSMUST00000032704.5 |
C230052I12Rik
|
RIKEN cDNA C230052I12 gene |
| chr15_-_11905609 | 2.08 |
ENSMUST00000066529.3
|
Npr3
|
natriuretic peptide receptor 3 |
| chr4_+_115088708 | 1.99 |
ENSMUST00000171877.1
ENSMUST00000177647.1 ENSMUST00000106548.2 ENSMUST00000030488.2 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr19_+_34100943 | 1.95 |
ENSMUST00000025685.6
|
Lipm
|
lipase, family member M |
| chr1_+_44551483 | 1.94 |
ENSMUST00000074525.3
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_-_30073554 | 1.93 |
ENSMUST00000026846.6
|
Tyms
|
thymidylate synthase |
| chr17_+_34593388 | 1.91 |
ENSMUST00000174532.1
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr15_-_100424092 | 1.89 |
ENSMUST00000154676.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr7_+_141447645 | 1.89 |
ENSMUST00000106004.1
ENSMUST00000106003.1 |
Rplp2
|
ribosomal protein, large P2 |
| chr2_-_148443543 | 1.84 |
ENSMUST00000099269.3
|
Cd93
|
CD93 antigen |
| chr11_-_45955465 | 1.84 |
ENSMUST00000011398.6
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr14_-_54994541 | 1.82 |
ENSMUST00000153783.1
ENSMUST00000168485.1 ENSMUST00000102803.3 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr1_+_165302625 | 1.78 |
ENSMUST00000111450.1
|
Gpr161
|
G protein-coupled receptor 161 |
| chr9_-_21037775 | 1.78 |
ENSMUST00000180870.1
|
Gm26592
|
predicted gene, 26592 |
| chr12_+_78861693 | 1.75 |
ENSMUST00000071230.7
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr9_-_109234754 | 1.70 |
ENSMUST00000079548.6
|
Fbxw20
|
F-box and WD-40 domain protein 20 |
| chrX_-_10216437 | 1.69 |
ENSMUST00000115534.1
|
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr7_+_131032061 | 1.66 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr5_-_100500592 | 1.65 |
ENSMUST00000149714.1
ENSMUST00000046154.5 |
Lin54
|
lin-54 homolog (C. elegans) |
| chr14_+_30479565 | 1.64 |
ENSMUST00000022535.7
|
Dcp1a
|
DCP1 decapping enzyme homolog A (S. cerevisiae) |
| chr8_-_35826435 | 1.63 |
ENSMUST00000060128.5
|
Cldn23
|
claudin 23 |
| chr18_+_65582390 | 1.56 |
ENSMUST00000169679.1
ENSMUST00000183326.1 |
Zfp532
|
zinc finger protein 532 |
| chr19_+_29367447 | 1.55 |
ENSMUST00000016640.7
|
Cd274
|
CD274 antigen |
| chr17_+_7170101 | 1.54 |
ENSMUST00000024575.6
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr11_+_3289880 | 1.50 |
ENSMUST00000110043.1
ENSMUST00000094471.3 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr11_-_45955183 | 1.50 |
ENSMUST00000109254.1
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr6_-_57844493 | 1.49 |
ENSMUST00000081186.3
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr3_+_92365178 | 1.49 |
ENSMUST00000050397.1
|
Sprr2f
|
small proline-rich protein 2F |
| chr18_+_65582239 | 1.47 |
ENSMUST00000182684.1
|
Zfp532
|
zinc finger protein 532 |
| chrX_+_7841800 | 1.46 |
ENSMUST00000033494.9
ENSMUST00000115666.1 |
Otud5
|
OTU domain containing 5 |
| chr2_+_16356744 | 1.46 |
ENSMUST00000114703.3
|
Plxdc2
|
plexin domain containing 2 |
| chr19_-_55241236 | 1.43 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr7_+_113514085 | 1.39 |
ENSMUST00000122890.1
|
Far1
|
fatty acyl CoA reductase 1 |
| chrX_+_74254736 | 1.34 |
ENSMUST00000096424.4
|
Emd
|
emerin |
| chr6_-_92244645 | 1.33 |
ENSMUST00000006046.4
|
Trh
|
thyrotropin releasing hormone |
| chr11_-_96111962 | 1.32 |
ENSMUST00000097162.5
ENSMUST00000068686.6 |
Calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr7_-_126200413 | 1.31 |
ENSMUST00000163959.1
|
Xpo6
|
exportin 6 |
| chrX_-_7572843 | 1.30 |
ENSMUST00000132788.1
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr5_-_110343009 | 1.29 |
ENSMUST00000058016.9
ENSMUST00000112478.3 |
P2rx2
|
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr7_+_35397046 | 1.28 |
ENSMUST00000079414.5
|
Cep89
|
centrosomal protein 89 |
| chr7_-_126594941 | 1.28 |
ENSMUST00000058429.5
|
Il27
|
interleukin 27 |
| chr19_+_5568002 | 1.21 |
ENSMUST00000096318.3
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr2_+_127854628 | 1.20 |
ENSMUST00000028859.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chrX_+_56346390 | 1.19 |
ENSMUST00000101560.3
|
Zfp449
|
zinc finger protein 449 |
| chr1_-_162740540 | 1.17 |
ENSMUST00000028016.9
ENSMUST00000182660.1 |
Prrc2c
|
proline-rich coiled-coil 2C |
| chr9_-_99358518 | 1.15 |
ENSMUST00000042158.6
|
Esyt3
|
extended synaptotagmin-like protein 3 |
| chr4_-_153482768 | 1.14 |
ENSMUST00000105646.2
|
Ajap1
|
adherens junction associated protein 1 |
| chr10_-_130127055 | 1.11 |
ENSMUST00000074161.1
|
Olfr824
|
olfactory receptor 824 |
| chr11_-_78751656 | 1.09 |
ENSMUST00000059468.4
|
Fam58b
|
family with sequence similarity 58, member B |
| chr14_-_79481268 | 1.05 |
ENSMUST00000022601.5
|
Wbp4
|
WW domain binding protein 4 |
| chrX_+_138914422 | 1.04 |
ENSMUST00000064937.7
ENSMUST00000113052.1 |
Nrk
|
Nik related kinase |
| chr9_+_35423582 | 1.03 |
ENSMUST00000154652.1
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr8_-_60954726 | 1.03 |
ENSMUST00000110302.1
|
Clcn3
|
chloride channel 3 |
| chr6_-_116716888 | 1.01 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr5_+_145114280 | 1.00 |
ENSMUST00000141602.1
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr1_-_85109846 | 0.99 |
ENSMUST00000161675.1
ENSMUST00000160792.1 |
A530032D15Rik
|
RIKEN cDNA A530032D15Rik gene |
| chr3_-_41742471 | 0.99 |
ENSMUST00000026866.8
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr15_-_75909319 | 0.99 |
ENSMUST00000089680.3
ENSMUST00000141268.1 ENSMUST00000023235.6 ENSMUST00000109972.2 ENSMUST00000089681.5 ENSMUST00000109975.3 ENSMUST00000154584.1 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_-_48594373 | 0.99 |
ENSMUST00000088898.4
ENSMUST00000072292.5 |
Zfp280c
|
zinc finger protein 280C |
| chr15_-_75909289 | 0.96 |
ENSMUST00000145764.1
ENSMUST00000116440.2 ENSMUST00000151066.1 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_+_74254679 | 0.96 |
ENSMUST00000002029.6
|
Emd
|
emerin |
| chr2_-_154769932 | 0.94 |
ENSMUST00000184654.1
|
Gm14214
|
predicted gene 14214 |
| chr17_+_34972687 | 0.92 |
ENSMUST00000007248.3
|
Hspa1l
|
heat shock protein 1-like |
| chrX_+_7842056 | 0.89 |
ENSMUST00000115667.3
ENSMUST00000115668.3 ENSMUST00000115665.1 |
Otud5
|
OTU domain containing 5 |
| chr3_+_84952146 | 0.89 |
ENSMUST00000029727.7
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr15_-_85578070 | 0.88 |
ENSMUST00000109424.2
|
Wnt7b
|
wingless-related MMTV integration site 7B |
| chr2_+_163225363 | 0.87 |
ENSMUST00000099110.3
ENSMUST00000165937.1 |
Tox2
|
TOX high mobility group box family member 2 |
| chr6_-_34317442 | 0.86 |
ENSMUST00000154655.1
ENSMUST00000102980.4 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr8_+_33599608 | 0.86 |
ENSMUST00000009774.9
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| chr17_-_65884902 | 0.85 |
ENSMUST00000024905.9
|
Ralbp1
|
ralA binding protein 1 |
| chr2_-_25983056 | 0.83 |
ENSMUST00000127823.1
ENSMUST00000134882.1 |
Camsap1
|
calmodulin regulated spectrin-associated protein 1 |
| chr11_-_70969953 | 0.82 |
ENSMUST00000108530.1
ENSMUST00000035283.4 ENSMUST00000108531.1 |
Nup88
|
nucleoporin 88 |
| chrX_+_52912232 | 0.81 |
ENSMUST00000078944.6
ENSMUST00000101587.3 ENSMUST00000154864.2 |
Phf6
|
PHD finger protein 6 |
| chrX_+_74254782 | 0.81 |
ENSMUST00000119197.1
ENSMUST00000088313.4 |
Emd
|
emerin |
| chr7_-_126200474 | 0.80 |
ENSMUST00000168189.1
|
Xpo6
|
exportin 6 |
| chr11_-_82829024 | 0.79 |
ENSMUST00000021036.6
ENSMUST00000074515.4 ENSMUST00000103218.2 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr19_-_45749563 | 0.78 |
ENSMUST00000070215.7
|
Npm3
|
nucleoplasmin 3 |
| chr4_-_132422394 | 0.74 |
ENSMUST00000152271.1
ENSMUST00000084170.5 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr18_+_65582281 | 0.70 |
ENSMUST00000183319.1
|
Zfp532
|
zinc finger protein 532 |
| chr15_+_54745702 | 0.69 |
ENSMUST00000050027.8
|
Nov
|
nephroblastoma overexpressed gene |
| chr11_+_102885160 | 0.69 |
ENSMUST00000100369.3
|
Fam187a
|
family with sequence similarity 187, member A |
| chr9_+_32372409 | 0.65 |
ENSMUST00000047334.8
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr19_+_6909692 | 0.62 |
ENSMUST00000088257.7
|
Trmt112
|
tRNA methyltransferase 11-2 |
| chr10_+_62449489 | 0.61 |
ENSMUST00000181110.1
|
4930507D05Rik
|
RIKEN cDNA 4930507D05 gene |
| chr16_+_44139821 | 0.61 |
ENSMUST00000159514.1
ENSMUST00000161326.1 ENSMUST00000063520.8 ENSMUST00000063542.7 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr4_-_139352538 | 0.61 |
ENSMUST00000102503.3
|
Mrto4
|
MRT4, mRNA turnover 4, homolog (S. cerevisiae) |
| chr4_-_141598206 | 0.60 |
ENSMUST00000131317.1
ENSMUST00000006381.4 ENSMUST00000129602.1 |
Fblim1
|
filamin binding LIM protein 1 |
| chr9_-_109626059 | 0.60 |
ENSMUST00000073962.6
|
Fbxw24
|
F-box and WD-40 domain protein 24 |
| chr8_+_11556061 | 0.60 |
ENSMUST00000054399.4
|
Ing1
|
inhibitor of growth family, member 1 |
| chr7_+_101969796 | 0.60 |
ENSMUST00000084852.5
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr8_+_82863351 | 0.57 |
ENSMUST00000078525.5
|
Rnf150
|
ring finger protein 150 |
| chr11_+_22990519 | 0.57 |
ENSMUST00000173867.1
ENSMUST00000020562.4 |
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 4.2 | 12.6 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 3.5 | 10.4 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 3.3 | 9.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 3.1 | 12.3 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 2.8 | 13.9 | GO:0071104 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 2.4 | 7.2 | GO:2000451 | immune complex clearance by monocytes and macrophages(GO:0002436) astrocyte chemotaxis(GO:0035700) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 2.3 | 16.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 2.3 | 6.8 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) histone H3-S10 phosphorylation(GO:0043987) |
| 2.1 | 16.6 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 1.9 | 11.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.9 | 13.0 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.8 | 10.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.6 | 4.8 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 1.3 | 3.9 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) positive regulation of actin filament-based movement(GO:1903116) |
| 1.2 | 3.7 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.1 | 5.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.9 | 3.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.8 | 4.1 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.8 | 4.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.8 | 3.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.7 | 12.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.7 | 5.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.7 | 7.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.6 | 5.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.6 | 3.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 2.4 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.6 | 2.3 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) positive regulation of response to alcohol(GO:1901421) |
| 0.6 | 2.8 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.5 | 2.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.5 | 2.9 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.5 | 10.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.5 | 2.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.5 | 1.9 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.5 | 6.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 5.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 1.7 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.4 | 10.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.4 | 1.8 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.0 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.3 | 1.0 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.3 | 1.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 | 5.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 3.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 1.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 1.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 2.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.3 | 1.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.3 | 3.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.3 | 1.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 2.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 2.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 2.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 1.4 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 0.7 | GO:1990523 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) negative regulation of sensory perception of pain(GO:1904057) bone regeneration(GO:1990523) |
| 0.2 | 0.6 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.2 | 1.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 3.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 0.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 2.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 3.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 3.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 1.6 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 1.5 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.2 | 6.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.2 | 1.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 1.3 | GO:0030432 | peristalsis(GO:0030432) |
| 0.2 | 2.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 0.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 9.4 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.1 | 1.0 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 2.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 1.3 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.1 | 4.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 2.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 4.1 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 1.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 4.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 1.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 3.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.3 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 4.1 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.5 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 0.4 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.1 | 4.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 2.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 5.7 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.0 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.3 | GO:1904871 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 1.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 10.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 2.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.9 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.3 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 1.0 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 2.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.6 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.7 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.6 | 9.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.3 | 6.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.0 | 7.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.9 | 4.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.8 | 3.3 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.8 | 4.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.7 | 3.7 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.6 | 3.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.6 | 5.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 12.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 4.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.4 | 1.7 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.4 | 2.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 3.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 3.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 2.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 10.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 5.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 3.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 2.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.6 | GO:0036449 | cortical microtubule cytoskeleton(GO:0030981) microtubule minus-end(GO:0036449) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 5.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.6 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 2.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 9.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 1.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.8 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 5.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 13.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 7.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 1.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 3.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 35.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 5.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 5.6 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 10.7 | GO:0030054 | cell junction(GO:0030054) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0004349 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 3.3 | 26.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 2.4 | 7.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 1.7 | 6.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.6 | 11.3 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 1.4 | 16.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 1.3 | 6.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.1 | 3.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.9 | 13.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.8 | 4.1 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.8 | 2.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.8 | 12.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.8 | 2.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.7 | 11.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 3.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 9.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.5 | 1.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.5 | 2.5 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.5 | 4.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.5 | 4.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.5 | 5.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 2.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 3.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.3 | 2.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 2.4 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 4.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 1.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 2.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 2.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 2.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 12.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 12.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 0.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 5.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 3.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 2.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 1.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 2.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 10.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 10.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 2.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 2.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 5.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.3 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 2.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 3.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 8.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 21.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 3.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 11.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.3 | 4.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 10.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 10.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 10.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 2.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 12.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 15.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 9.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 4.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 1.0 | 16.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.9 | 15.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.7 | 8.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.6 | 8.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.5 | 12.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 16.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 9.8 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.3 | 12.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 3.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 9.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 4.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 3.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 9.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.7 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 2.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 5.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 8.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 3.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.9 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 3.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 13.8 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 4.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |