Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tlx2
Z-value: 0.63
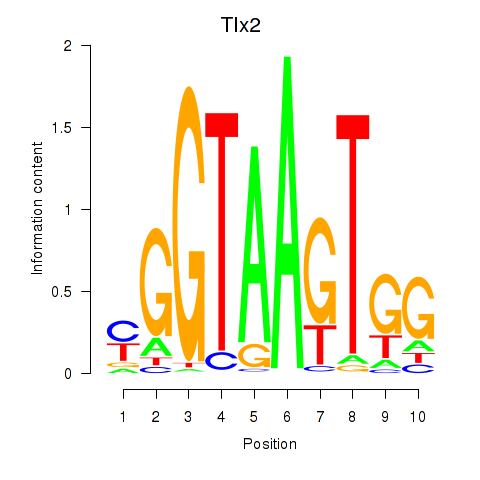
Transcription factors associated with Tlx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tlx2
|
ENSMUSG00000068327.3 | T cell leukemia, homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tlx2 | mm10_v2_chr6_-_83070225_83070377 | -0.20 | 2.4e-01 | Click! |
Activity profile of Tlx2 motif
Sorted Z-values of Tlx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_105933832 | 2.41 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr7_-_30924169 | 2.18 |
ENSMUST00000074671.6
|
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr1_-_150466165 | 1.98 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr8_-_111691002 | 1.94 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr3_-_98630309 | 1.89 |
ENSMUST00000044094.4
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr17_-_31144271 | 1.88 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr8_+_105048592 | 1.74 |
ENSMUST00000093222.5
ENSMUST00000093223.3 |
Ces3a
|
carboxylesterase 3A |
| chr17_-_84682932 | 1.63 |
ENSMUST00000066175.3
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr15_+_77729091 | 1.58 |
ENSMUST00000109775.2
|
Apol9b
|
apolipoprotein L 9b |
| chr5_+_137981512 | 1.48 |
ENSMUST00000035390.5
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr4_-_60499332 | 1.47 |
ENSMUST00000135953.1
|
Mup1
|
major urinary protein 1 |
| chr19_-_7802578 | 1.46 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr17_+_84683113 | 1.39 |
ENSMUST00000045714.8
|
Abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr1_-_162898665 | 1.38 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr17_+_84683131 | 1.37 |
ENSMUST00000171915.1
|
Abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr4_+_63362443 | 1.36 |
ENSMUST00000075341.3
|
Orm2
|
orosomucoid 2 |
| chr18_+_60376029 | 1.31 |
ENSMUST00000066912.5
ENSMUST00000032473.6 |
Iigp1
|
interferon inducible GTPase 1 |
| chr15_-_100687908 | 1.31 |
ENSMUST00000023775.7
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr13_-_55426783 | 1.29 |
ENSMUST00000021948.8
|
F12
|
coagulation factor XII (Hageman factor) |
| chr5_-_145879857 | 1.21 |
ENSMUST00000035918.7
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr9_-_71168657 | 1.18 |
ENSMUST00000113570.1
|
Aqp9
|
aquaporin 9 |
| chr11_+_115462464 | 1.15 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr9_+_111439063 | 1.11 |
ENSMUST00000111879.3
|
Dclk3
|
doublecortin-like kinase 3 |
| chr5_-_105343929 | 1.04 |
ENSMUST00000183149.1
|
Gbp11
|
guanylate binding protein 11 |
| chr13_+_23807027 | 1.03 |
ENSMUST00000006786.4
ENSMUST00000099697.2 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr1_-_139858684 | 1.03 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr3_-_98753465 | 0.98 |
ENSMUST00000094050.4
ENSMUST00000090743.6 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr4_+_63356152 | 0.97 |
ENSMUST00000006687.4
|
Orm3
|
orosomucoid 3 |
| chr9_-_48605147 | 0.96 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr6_-_21851914 | 0.94 |
ENSMUST00000134635.1
ENSMUST00000123116.1 ENSMUST00000120965.1 |
Tspan12
|
tetraspanin 12 |
| chr13_-_55426769 | 0.93 |
ENSMUST00000170921.1
|
F12
|
coagulation factor XII (Hageman factor) |
| chr2_+_25054396 | 0.88 |
ENSMUST00000102931.4
ENSMUST00000074422.7 ENSMUST00000132172.1 ENSMUST00000114388.1 ENSMUST00000114386.1 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr4_-_104876383 | 0.88 |
ENSMUST00000064873.8
ENSMUST00000106808.3 ENSMUST00000048947.8 |
C8a
|
complement component 8, alpha polypeptide |
| chr2_+_25054355 | 0.88 |
ENSMUST00000100334.4
ENSMUST00000152122.1 ENSMUST00000116574.3 ENSMUST00000006646.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr3_-_113574758 | 0.87 |
ENSMUST00000106540.1
|
Amy1
|
amylase 1, salivary |
| chr18_+_20247340 | 0.87 |
ENSMUST00000054128.6
|
Dsg1c
|
desmoglein 1 gamma |
| chr9_+_121642716 | 0.87 |
ENSMUST00000035115.4
|
Vipr1
|
vasoactive intestinal peptide receptor 1 |
| chr4_-_141846277 | 0.83 |
ENSMUST00000105781.1
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr13_+_19342154 | 0.82 |
ENSMUST00000103566.3
|
Tcrg-C4
|
T cell receptor gamma, constant 4 |
| chr4_-_49506538 | 0.82 |
ENSMUST00000043056.2
|
Baat
|
bile acid-Coenzyme A: amino acid N-acyltransferase |
| chrX_+_20549780 | 0.81 |
ENSMUST00000023832.6
|
Rgn
|
regucalcin |
| chr19_-_11266122 | 0.80 |
ENSMUST00000169159.1
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr18_-_3337467 | 0.80 |
ENSMUST00000154135.1
|
Crem
|
cAMP responsive element modulator |
| chr6_-_124636085 | 0.79 |
ENSMUST00000068797.2
|
Gm5077
|
predicted gene 5077 |
| chr4_-_141846359 | 0.78 |
ENSMUST00000037059.10
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr4_-_108071327 | 0.76 |
ENSMUST00000106701.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr9_-_103219823 | 0.75 |
ENSMUST00000168142.1
|
Trf
|
transferrin |
| chr12_-_98577940 | 0.75 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr8_+_70083509 | 0.72 |
ENSMUST00000007738.9
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chrX_-_139782353 | 0.72 |
ENSMUST00000101217.3
|
Ripply1
|
ripply1 homolog (zebrafish) |
| chr10_-_115362191 | 0.70 |
ENSMUST00000092170.5
|
Tmem19
|
transmembrane protein 19 |
| chr3_-_113574242 | 0.70 |
ENSMUST00000142505.2
|
Amy1
|
amylase 1, salivary |
| chr18_-_34720269 | 0.70 |
ENSMUST00000025224.7
|
Gfra3
|
glial cell line derived neurotrophic factor family receptor alpha 3 |
| chr2_+_23069057 | 0.67 |
ENSMUST00000114526.1
ENSMUST00000114529.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr1_-_182409020 | 0.65 |
ENSMUST00000097444.1
|
Gm10517
|
predicted gene 10517 |
| chr6_+_122513583 | 0.65 |
ENSMUST00000032210.7
ENSMUST00000148517.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr11_+_51763682 | 0.65 |
ENSMUST00000020653.5
|
Sar1b
|
SAR1 gene homolog B (S. cerevisiae) |
| chr18_-_3337614 | 0.64 |
ENSMUST00000150235.1
ENSMUST00000154470.1 |
Crem
|
cAMP responsive element modulator |
| chr14_-_61360395 | 0.64 |
ENSMUST00000022494.8
|
Ebpl
|
emopamil binding protein-like |
| chr1_+_132298606 | 0.64 |
ENSMUST00000046071.4
|
Klhdc8a
|
kelch domain containing 8A |
| chr14_+_52824340 | 0.64 |
ENSMUST00000103648.2
|
Trav11d
|
T cell receptor alpha variable 11D |
| chr15_+_75596645 | 0.63 |
ENSMUST00000023243.4
|
Gpihbp1
|
GPI-anchored HDL-binding protein 1 |
| chr11_+_102041509 | 0.62 |
ENSMUST00000123895.1
ENSMUST00000017453.5 ENSMUST00000107163.2 ENSMUST00000107164.2 |
Cd300lg
|
CD300 antigen like family member G |
| chr7_+_79273201 | 0.61 |
ENSMUST00000037315.6
|
Abhd2
|
abhydrolase domain containing 2 |
| chr2_+_23069210 | 0.61 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr18_-_3281036 | 0.60 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr16_+_38902305 | 0.60 |
ENSMUST00000023478.7
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr11_+_102836296 | 0.60 |
ENSMUST00000021302.8
ENSMUST00000107072.1 |
Higd1b
|
HIG1 domain family, member 1B |
| chr2_-_25493432 | 0.59 |
ENSMUST00000114245.1
|
Lcn12
|
lipocalin 12 |
| chr9_-_21989427 | 0.59 |
ENSMUST00000045726.6
|
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr18_-_3337539 | 0.58 |
ENSMUST00000142690.1
ENSMUST00000025069.4 ENSMUST00000082141.5 ENSMUST00000165086.1 ENSMUST00000149803.1 |
Crem
|
cAMP responsive element modulator |
| chr13_-_56548534 | 0.57 |
ENSMUST00000062806.4
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr12_+_100779088 | 0.57 |
ENSMUST00000110069.1
|
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr12_+_100779074 | 0.56 |
ENSMUST00000110073.1
ENSMUST00000110070.1 |
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr2_-_172458596 | 0.56 |
ENSMUST00000099060.1
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr17_-_36058371 | 0.56 |
ENSMUST00000113742.2
|
Gm11127
|
predicted gene 11127 |
| chr16_+_20733104 | 0.54 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr19_+_45076105 | 0.54 |
ENSMUST00000026234.4
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr9_-_51328898 | 0.53 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr17_+_43667389 | 0.53 |
ENSMUST00000170988.1
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr7_-_30598863 | 0.53 |
ENSMUST00000108150.1
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr8_+_13037802 | 0.51 |
ENSMUST00000152034.1
ENSMUST00000128418.1 |
F10
|
coagulation factor X |
| chrX_+_10252305 | 0.51 |
ENSMUST00000049910.6
|
Otc
|
ornithine transcarbamylase |
| chr10_+_127849917 | 0.50 |
ENSMUST00000077530.2
|
Rdh19
|
retinol dehydrogenase 19 |
| chr11_-_21572193 | 0.50 |
ENSMUST00000102874.4
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr6_-_35326123 | 0.50 |
ENSMUST00000051176.7
|
Fam180a
|
family with sequence similarity 180, member A |
| chr9_-_21927515 | 0.50 |
ENSMUST00000178988.1
ENSMUST00000046831.9 |
Tmem205
|
transmembrane protein 205 |
| chrX_+_10252361 | 0.48 |
ENSMUST00000115528.2
|
Otc
|
ornithine transcarbamylase |
| chr3_+_121723515 | 0.48 |
ENSMUST00000029771.8
|
F3
|
coagulation factor III |
| chr16_-_18413452 | 0.47 |
ENSMUST00000165430.1
ENSMUST00000147720.1 |
Comt
|
catechol-O-methyltransferase |
| chr18_-_60273267 | 0.47 |
ENSMUST00000090260.4
|
Gm4841
|
predicted gene 4841 |
| chr9_+_108080436 | 0.45 |
ENSMUST00000035211.7
ENSMUST00000162886.1 |
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr1_-_132139605 | 0.45 |
ENSMUST00000112362.2
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr2_+_30364262 | 0.45 |
ENSMUST00000142801.1
ENSMUST00000100214.3 |
Fam73b
|
family with sequence similarity 73, member B |
| chr9_-_50728067 | 0.45 |
ENSMUST00000117646.1
|
Dixdc1
|
DIX domain containing 1 |
| chr9_-_50727921 | 0.45 |
ENSMUST00000118707.1
ENSMUST00000034566.8 |
Dixdc1
|
DIX domain containing 1 |
| chrX_+_141475385 | 0.44 |
ENSMUST00000112931.1
ENSMUST00000112930.1 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr17_+_35439155 | 0.44 |
ENSMUST00000071951.6
ENSMUST00000078205.7 ENSMUST00000116598.3 ENSMUST00000076256.7 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr12_+_41024329 | 0.44 |
ENSMUST00000134965.1
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr7_+_131410601 | 0.44 |
ENSMUST00000015829.7
ENSMUST00000117518.1 |
Acadsb
|
acyl-Coenzyme A dehydrogenase, short/branched chain |
| chr10_+_18845071 | 0.43 |
ENSMUST00000019998.7
|
Perp
|
PERP, TP53 apoptosis effector |
| chr6_-_23839137 | 0.43 |
ENSMUST00000166458.2
ENSMUST00000142913.2 ENSMUST00000115357.1 ENSMUST00000069074.7 ENSMUST00000115361.2 ENSMUST00000018122.7 ENSMUST00000115355.1 ENSMUST00000115356.2 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr14_-_66868572 | 0.43 |
ENSMUST00000022629.8
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr10_+_13966268 | 0.43 |
ENSMUST00000015645.4
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr17_+_34263209 | 0.43 |
ENSMUST00000040828.5
|
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr11_-_4160286 | 0.42 |
ENSMUST00000093381.4
ENSMUST00000101626.2 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr9_+_44107268 | 0.42 |
ENSMUST00000114821.2
ENSMUST00000114818.2 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr4_+_134396320 | 0.42 |
ENSMUST00000105869.2
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr19_+_36554661 | 0.42 |
ENSMUST00000169036.2
ENSMUST00000047247.5 |
Hectd2
|
HECT domain containing 2 |
| chr15_+_54745702 | 0.41 |
ENSMUST00000050027.8
|
Nov
|
nephroblastoma overexpressed gene |
| chr10_+_116301374 | 0.41 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr3_-_101604580 | 0.41 |
ENSMUST00000036493.6
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr7_-_48456331 | 0.41 |
ENSMUST00000094384.3
|
Mrgprb1
|
MAS-related GPR, member B1 |
| chr13_-_36734450 | 0.40 |
ENSMUST00000037623.8
|
Nrn1
|
neuritin 1 |
| chr12_+_41024090 | 0.40 |
ENSMUST00000132121.1
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr2_+_34772089 | 0.40 |
ENSMUST00000028222.6
ENSMUST00000100171.2 |
Hspa5
|
heat shock protein 5 |
| chr9_+_44107286 | 0.40 |
ENSMUST00000152956.1
ENSMUST00000114815.1 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr6_+_117168535 | 0.40 |
ENSMUST00000112866.1
ENSMUST00000112871.1 ENSMUST00000073043.4 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr16_+_43363855 | 0.39 |
ENSMUST00000156367.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_-_45204083 | 0.39 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr1_-_162859684 | 0.39 |
ENSMUST00000131058.1
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr6_+_122513643 | 0.39 |
ENSMUST00000118626.1
|
Mfap5
|
microfibrillar associated protein 5 |
| chr4_-_41517326 | 0.39 |
ENSMUST00000030152.6
ENSMUST00000095126.4 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr16_+_22892035 | 0.38 |
ENSMUST00000023583.5
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr18_+_60293372 | 0.38 |
ENSMUST00000171297.1
|
F830016B08Rik
|
RIKEN cDNA F830016B08 gene |
| chr2_+_30364227 | 0.38 |
ENSMUST00000077977.7
ENSMUST00000140075.2 |
Fam73b
|
family with sequence similarity 73, member B |
| chr14_+_32028989 | 0.38 |
ENSMUST00000022460.4
|
Galnt15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr10_-_127121125 | 0.38 |
ENSMUST00000164259.1
ENSMUST00000080975.4 |
Os9
|
amplified in osteosarcoma |
| chr2_-_150362708 | 0.38 |
ENSMUST00000051153.5
|
3300002I08Rik
|
RIKEN cDNA 3300002I08 gene |
| chr9_-_107710475 | 0.37 |
ENSMUST00000080560.3
|
Sema3f
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr16_-_45408955 | 0.37 |
ENSMUST00000163230.1
|
Cd200
|
CD200 antigen |
| chr9_-_29412204 | 0.37 |
ENSMUST00000115237.1
|
Ntm
|
neurotrimin |
| chr16_-_23029062 | 0.37 |
ENSMUST00000115349.2
|
Kng2
|
kininogen 2 |
| chr8_+_94472763 | 0.37 |
ENSMUST00000053085.5
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr10_+_79890853 | 0.37 |
ENSMUST00000061653.7
|
Cfd
|
complement factor D (adipsin) |
| chr10_-_86705485 | 0.37 |
ENSMUST00000020238.7
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr8_+_46492789 | 0.36 |
ENSMUST00000110371.1
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr18_+_56432116 | 0.36 |
ENSMUST00000070166.5
|
Gramd3
|
GRAM domain containing 3 |
| chr17_+_24850515 | 0.36 |
ENSMUST00000154363.1
ENSMUST00000169200.1 |
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr1_-_106759727 | 0.36 |
ENSMUST00000010049.4
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr2_-_168741898 | 0.36 |
ENSMUST00000109176.1
ENSMUST00000178504.1 |
Atp9a
|
ATPase, class II, type 9A |
| chr15_+_32920723 | 0.36 |
ENSMUST00000022871.5
|
Sdc2
|
syndecan 2 |
| chr11_+_6658510 | 0.35 |
ENSMUST00000045374.7
|
Ramp3
|
receptor (calcitonin) activity modifying protein 3 |
| chr2_-_69342600 | 0.35 |
ENSMUST00000102709.1
ENSMUST00000102710.3 ENSMUST00000180142.1 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr9_+_21927471 | 0.35 |
ENSMUST00000170304.1
ENSMUST00000006403.6 |
Ccdc159
|
coiled-coil domain containing 159 |
| chr12_-_44210061 | 0.35 |
ENSMUST00000015049.3
|
Dnajb9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr16_-_23029080 | 0.35 |
ENSMUST00000100046.2
|
Kng2
|
kininogen 2 |
| chr18_+_20376723 | 0.34 |
ENSMUST00000076737.6
|
Dsg1b
|
desmoglein 1 beta |
| chr16_-_23029012 | 0.34 |
ENSMUST00000039338.6
|
Kng2
|
kininogen 2 |
| chr2_+_177768044 | 0.34 |
ENSMUST00000108942.3
|
Gm14322
|
predicted gene 14322 |
| chr5_-_86518578 | 0.34 |
ENSMUST00000134179.1
|
Tmprss11g
|
transmembrane protease, serine 11g |
| chr2_+_4717825 | 0.34 |
ENSMUST00000184139.1
ENSMUST00000115022.1 |
Bend7
|
BEN domain containing 7 |
| chr10_+_4710119 | 0.33 |
ENSMUST00000105588.1
ENSMUST00000105589.1 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr7_+_45617575 | 0.33 |
ENSMUST00000008605.5
|
Fut1
|
fucosyltransferase 1 |
| chr1_+_16688405 | 0.33 |
ENSMUST00000026881.4
|
Ly96
|
lymphocyte antigen 96 |
| chr17_-_56109775 | 0.33 |
ENSMUST00000002908.7
|
Plin4
|
perilipin 4 |
| chr4_-_120747248 | 0.33 |
ENSMUST00000030376.7
|
Kcnq4
|
potassium voltage-gated channel, subfamily Q, member 4 |
| chr7_-_44306903 | 0.32 |
ENSMUST00000004587.9
|
Clec11a
|
C-type lectin domain family 11, member a |
| chr19_+_4003334 | 0.32 |
ENSMUST00000025806.3
|
Doc2g
|
double C2, gamma |
| chr2_-_163419508 | 0.32 |
ENSMUST00000046908.3
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr11_-_110251736 | 0.32 |
ENSMUST00000044003.7
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr2_-_130424242 | 0.32 |
ENSMUST00000089581.4
|
Pced1a
|
PC-esterase domain containing 1A |
| chr8_-_61760067 | 0.32 |
ENSMUST00000121493.1
|
Palld
|
palladin, cytoskeletal associated protein |
| chr5_+_28071356 | 0.32 |
ENSMUST00000059155.10
|
Insig1
|
insulin induced gene 1 |
| chr11_+_43474276 | 0.32 |
ENSMUST00000173002.1
ENSMUST00000057679.3 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr5_+_114146525 | 0.31 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr12_+_44269145 | 0.31 |
ENSMUST00000043082.8
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr17_-_73950172 | 0.31 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr10_+_94036001 | 0.31 |
ENSMUST00000020208.4
|
Fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr9_-_58313189 | 0.31 |
ENSMUST00000061799.8
|
Loxl1
|
lysyl oxidase-like 1 |
| chr9_-_71771535 | 0.30 |
ENSMUST00000122065.1
ENSMUST00000121322.1 ENSMUST00000072899.2 |
Cgnl1
|
cingulin-like 1 |
| chr19_-_24031006 | 0.30 |
ENSMUST00000096164.4
|
Fam189a2
|
family with sequence similarity 189, member A2 |
| chr9_+_44107226 | 0.30 |
ENSMUST00000114816.1
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr17_+_24850484 | 0.30 |
ENSMUST00000118788.1
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr3_-_151749877 | 0.30 |
ENSMUST00000029671.7
|
Ifi44
|
interferon-induced protein 44 |
| chr17_+_34240015 | 0.30 |
ENSMUST00000173764.1
|
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr9_+_124121534 | 0.30 |
ENSMUST00000111442.1
ENSMUST00000171499.2 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr5_+_22550391 | 0.29 |
ENSMUST00000181374.1
ENSMUST00000181764.1 ENSMUST00000181209.1 |
6030443J06Rik
|
RIKEN cDNA 6030443J06 gene |
| chr11_+_51650954 | 0.29 |
ENSMUST00000117859.1
ENSMUST00000064493.5 |
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr5_-_123141067 | 0.29 |
ENSMUST00000162697.1
ENSMUST00000160321.1 ENSMUST00000159637.1 |
AI480526
|
expressed sequence AI480526 |
| chr1_+_4808237 | 0.29 |
ENSMUST00000131119.1
|
Lypla1
|
lysophospholipase 1 |
| chr7_+_130774069 | 0.29 |
ENSMUST00000048453.5
|
Btbd16
|
BTB (POZ) domain containing 16 |
| chr2_-_30205772 | 0.29 |
ENSMUST00000113662.1
|
Ccbl1
|
cysteine conjugate-beta lyase 1 |
| chr6_-_119330723 | 0.29 |
ENSMUST00000068351.7
|
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr6_-_119330668 | 0.29 |
ENSMUST00000112756.1
|
Lrtm2
|
leucine-rich repeats and transmembrane domains 2 |
| chr18_+_20310738 | 0.29 |
ENSMUST00000077146.3
|
Dsg1a
|
desmoglein 1 alpha |
| chr2_-_30205794 | 0.28 |
ENSMUST00000113663.2
ENSMUST00000044038.3 |
Ccbl1
|
cysteine conjugate-beta lyase 1 |
| chr14_-_33447142 | 0.28 |
ENSMUST00000111944.3
ENSMUST00000022504.5 ENSMUST00000111945.2 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chr7_-_126897424 | 0.28 |
ENSMUST00000120007.1
|
Tmem219
|
transmembrane protein 219 |
| chr15_+_54571358 | 0.28 |
ENSMUST00000025356.2
|
Mal2
|
mal, T cell differentiation protein 2 |
| chr14_+_65837302 | 0.28 |
ENSMUST00000022614.5
|
Ccdc25
|
coiled-coil domain containing 25 |
| chr7_+_25619404 | 0.28 |
ENSMUST00000077338.5
ENSMUST00000085953.3 |
Atp5sl
|
ATP5S-like |
| chr5_-_91402905 | 0.27 |
ENSMUST00000121044.2
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr5_-_62766153 | 0.27 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_+_50110186 | 0.27 |
ENSMUST00000166318.1
ENSMUST00000036236.8 ENSMUST00000036225.8 |
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr17_+_24850742 | 0.27 |
ENSMUST00000149716.2
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr7_-_128740471 | 0.27 |
ENSMUST00000119081.1
ENSMUST00000057557.7 |
Mcmbp
|
MCM (minichromosome maintenance deficient) binding protein |
| chr17_+_24850654 | 0.27 |
ENSMUST00000130989.1
ENSMUST00000024974.9 |
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr4_-_155345696 | 0.27 |
ENSMUST00000103178.4
|
Prkcz
|
protein kinase C, zeta |
| chr7_-_45061706 | 0.27 |
ENSMUST00000107832.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tlx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0045796 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.9 | 2.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 | 2.2 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.4 | 1.7 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.3 | 1.0 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.3 | 1.2 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.3 | 0.8 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 1.3 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.2 | 1.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 2.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 1.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 0.8 | GO:1901896 | L-ascorbic acid biosynthetic process(GO:0019853) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 0.8 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.2 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.2 | 0.6 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 0.5 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.1 | 0.4 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.4 | GO:1990523 | negative regulation of sensory perception of pain(GO:1904057) bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 1.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.6 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 1.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.3 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.4 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.8 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.5 | GO:0090005 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 0.4 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.1 | 0.3 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 1.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 2.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.2 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.4 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.3 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 0.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.3 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.1 | 0.2 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 | 0.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.4 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.6 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.4 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 0.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 2.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.4 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.5 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.0 | 0.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.3 | GO:0031622 | positive regulation of fever generation(GO:0031622) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 2.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 1.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.7 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.2 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 1.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.1 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.4 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 1.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.0 | GO:0006296 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:1903301 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.2 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.0 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.0 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.0 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.7 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 1.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.4 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.4 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) pheromone activity(GO:0005186) |
| 0.3 | 1.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 1.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 1.2 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.2 | 1.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.9 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 3.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 0.8 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.2 | 1.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 2.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 4.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 1.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 1.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.3 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.8 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 0.3 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.3 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 0.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.2 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.9 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.8 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 11.4 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.0 | 1.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.8 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.2 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |