Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
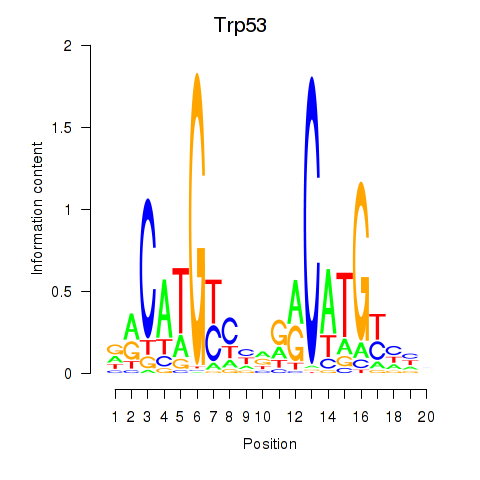
Results for Trp53
Z-value: 1.13
Transcription factors associated with Trp53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp53
|
ENSMUSG00000059552.7 | transformation related protein 53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp53 | mm10_v2_chr11_+_69580359_69580382 | 0.82 | 9.3e-10 | Click! |
Activity profile of Trp53 motif
Sorted Z-values of Trp53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_24760311 | 14.42 |
ENSMUST00000063956.5
|
Cd177
|
CD177 antigen |
| chr3_+_90669063 | 11.44 |
ENSMUST00000069927.8
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr16_+_36253046 | 9.17 |
ENSMUST00000063539.6
|
2010005H15Rik
|
RIKEN cDNA 2010005H15 gene |
| chr17_-_33890539 | 9.02 |
ENSMUST00000173386.1
|
Kifc1
|
kinesin family member C1 |
| chr17_-_33890584 | 8.88 |
ENSMUST00000114361.2
ENSMUST00000173492.1 |
Kifc1
|
kinesin family member C1 |
| chr17_-_35066170 | 7.76 |
ENSMUST00000174190.1
ENSMUST00000097337.1 |
AU023871
|
expressed sequence AU023871 |
| chr10_+_75564086 | 6.65 |
ENSMUST00000141062.1
ENSMUST00000152657.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr4_+_115057683 | 6.46 |
ENSMUST00000161601.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr4_+_115057410 | 6.23 |
ENSMUST00000136946.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr17_+_28207778 | 6.12 |
ENSMUST00000002327.5
|
Def6
|
differentially expressed in FDCP 6 |
| chr8_+_94977101 | 6.04 |
ENSMUST00000179619.1
|
Gpr56
|
G protein-coupled receptor 56 |
| chr6_-_116461024 | 5.70 |
ENSMUST00000164547.1
ENSMUST00000170186.1 |
Alox5
|
arachidonate 5-lipoxygenase |
| chr14_-_55944536 | 5.58 |
ENSMUST00000022834.6
|
Cma1
|
chymase 1, mast cell |
| chr4_+_155790439 | 5.36 |
ENSMUST00000165000.1
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr3_-_132950043 | 5.18 |
ENSMUST00000117164.1
ENSMUST00000093971.4 ENSMUST00000042729.9 ENSMUST00000042744.9 ENSMUST00000117811.1 |
Npnt
|
nephronectin |
| chr4_-_134254076 | 5.08 |
ENSMUST00000060050.5
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr8_-_22185758 | 4.97 |
ENSMUST00000046916.7
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr6_-_116461151 | 4.85 |
ENSMUST00000026795.6
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr11_-_102925086 | 4.78 |
ENSMUST00000021311.9
|
Kif18b
|
kinesin family member 18B |
| chr17_+_35821675 | 4.66 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr11_+_58948890 | 4.61 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chrX_-_142306170 | 4.11 |
ENSMUST00000134825.2
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr9_+_72438519 | 4.02 |
ENSMUST00000184604.1
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr15_+_85859689 | 3.90 |
ENSMUST00000170629.1
|
Gtse1
|
G two S phase expressed protein 1 |
| chr6_-_128891105 | 3.89 |
ENSMUST00000178918.1
ENSMUST00000160290.1 |
BC035044
|
cDNA sequence BC035044 |
| chr7_+_142498832 | 3.85 |
ENSMUST00000078497.8
ENSMUST00000105953.3 ENSMUST00000179658.1 ENSMUST00000105954.3 ENSMUST00000105952.3 ENSMUST00000105955.1 ENSMUST00000074187.6 ENSMUST00000180152.1 ENSMUST00000105950.4 ENSMUST00000105957.3 ENSMUST00000169299.2 ENSMUST00000105958.3 ENSMUST00000105949.1 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr9_+_72438534 | 3.81 |
ENSMUST00000034746.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr4_-_140810646 | 3.71 |
ENSMUST00000026377.2
|
Padi3
|
peptidyl arginine deiminase, type III |
| chr11_-_116076986 | 3.53 |
ENSMUST00000153408.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr2_-_69206146 | 3.45 |
ENSMUST00000127243.1
ENSMUST00000149643.1 ENSMUST00000167875.2 ENSMUST00000005365.8 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr4_+_155791172 | 3.32 |
ENSMUST00000105593.1
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr6_-_136941887 | 3.25 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr16_+_17144600 | 3.23 |
ENSMUST00000115702.1
|
Ydjc
|
YdjC homolog (bacterial) |
| chr17_+_26917091 | 3.20 |
ENSMUST00000078961.4
|
Kifc5b
|
kinesin family member C5B |
| chr14_-_37110087 | 3.16 |
ENSMUST00000179488.1
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr4_-_135573623 | 3.08 |
ENSMUST00000105855.1
|
Grhl3
|
grainyhead-like 3 (Drosophila) |
| chr9_+_45138437 | 2.94 |
ENSMUST00000060125.5
|
Scn4b
|
sodium channel, type IV, beta |
| chr9_-_21963568 | 2.92 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr8_-_88636117 | 2.82 |
ENSMUST00000034087.7
|
Snx20
|
sorting nexin 20 |
| chr12_-_78906929 | 2.82 |
ENSMUST00000021544.7
|
Plek2
|
pleckstrin 2 |
| chr11_-_102469839 | 2.79 |
ENSMUST00000103086.3
|
Itga2b
|
integrin alpha 2b |
| chr1_-_38664947 | 2.79 |
ENSMUST00000039827.7
ENSMUST00000027250.7 |
Aff3
|
AF4/FMR2 family, member 3 |
| chr2_-_69206133 | 2.69 |
ENSMUST00000112320.1
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr3_-_90389884 | 2.68 |
ENSMUST00000029541.5
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr6_-_136941494 | 2.64 |
ENSMUST00000111892.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr16_-_17144415 | 2.57 |
ENSMUST00000115709.1
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr6_-_136941694 | 2.45 |
ENSMUST00000032344.5
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr13_+_113317084 | 2.44 |
ENSMUST00000136755.2
|
BC067074
|
cDNA sequence BC067074 |
| chr14_-_54926784 | 2.38 |
ENSMUST00000022813.6
|
Efs
|
embryonal Fyn-associated substrate |
| chr5_+_144190284 | 2.36 |
ENSMUST00000060747.7
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr8_-_105851981 | 2.36 |
ENSMUST00000040776.4
|
Cenpt
|
centromere protein T |
| chr11_+_87760533 | 2.35 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr6_+_71707561 | 2.32 |
ENSMUST00000121469.1
|
Reep1
|
receptor accessory protein 1 |
| chr2_-_163397946 | 2.23 |
ENSMUST00000017961.4
ENSMUST00000109425.2 |
Jph2
|
junctophilin 2 |
| chr11_-_116077606 | 2.20 |
ENSMUST00000106450.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr1_+_171559186 | 2.19 |
ENSMUST00000004829.7
|
Cd244
|
CD244 natural killer cell receptor 2B4 |
| chr14_-_78725089 | 2.19 |
ENSMUST00000074729.5
|
Dgkh
|
diacylglycerol kinase, eta |
| chr17_+_29090969 | 2.13 |
ENSMUST00000119901.1
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr7_+_18718075 | 2.03 |
ENSMUST00000108481.1
ENSMUST00000051973.8 |
Psg22
|
pregnancy-specific glycoprotein 22 |
| chr7_+_103937382 | 2.00 |
ENSMUST00000098189.1
|
Olfr632
|
olfactory receptor 632 |
| chr16_+_38458887 | 1.99 |
ENSMUST00000099816.2
|
Cd80
|
CD80 antigen |
| chr11_-_106301801 | 1.97 |
ENSMUST00000103071.3
|
Gh
|
growth hormone |
| chr7_+_130865835 | 1.97 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr7_+_47050628 | 1.94 |
ENSMUST00000010451.5
|
Tmem86a
|
transmembrane protein 86A |
| chr5_+_34999111 | 1.94 |
ENSMUST00000114283.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_-_30073554 | 1.92 |
ENSMUST00000026846.6
|
Tyms
|
thymidylate synthase |
| chr5_+_34999046 | 1.91 |
ENSMUST00000114281.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr14_-_70175397 | 1.87 |
ENSMUST00000143393.1
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr9_-_121792478 | 1.79 |
ENSMUST00000035110.4
|
Hhatl
|
hedgehog acyltransferase-like |
| chr4_-_156059414 | 1.71 |
ENSMUST00000184348.1
|
Ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr5_+_34999070 | 1.70 |
ENSMUST00000114280.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr7_+_24884809 | 1.67 |
ENSMUST00000156372.1
ENSMUST00000124035.1 |
Rps19
|
ribosomal protein S19 |
| chr7_-_24545994 | 1.66 |
ENSMUST00000011776.6
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr7_+_130865756 | 1.61 |
ENSMUST00000120441.1
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr2_+_29889217 | 1.58 |
ENSMUST00000123335.1
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr8_+_68880491 | 1.55 |
ENSMUST00000015712.8
|
Lpl
|
lipoprotein lipase |
| chr5_+_136057267 | 1.54 |
ENSMUST00000006303.4
ENSMUST00000156530.1 |
Upk3bl
|
uroplakin 3B-like |
| chr17_+_78882003 | 1.51 |
ENSMUST00000180880.1
|
Gm26637
|
predicted gene, 26637 |
| chr10_+_21690845 | 1.50 |
ENSMUST00000071008.3
|
Gm5420
|
predicted gene 5420 |
| chr5_+_34989473 | 1.45 |
ENSMUST00000114284.1
ENSMUST00000114285.1 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr2_+_29890063 | 1.45 |
ENSMUST00000028128.6
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr3_+_96172327 | 1.39 |
ENSMUST00000076372.4
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr15_-_102350692 | 1.36 |
ENSMUST00000041208.7
|
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr9_-_120068263 | 1.35 |
ENSMUST00000064165.3
ENSMUST00000177637.1 |
Cx3cr1
|
chemokine (C-X3-C) receptor 1 |
| chr7_+_19851994 | 1.34 |
ENSMUST00000172815.1
|
Gm19345
|
predicted gene, 19345 |
| chr5_+_114707760 | 1.29 |
ENSMUST00000094441.4
|
Tchp
|
trichoplein, keratin filament binding |
| chr11_-_3527916 | 1.23 |
ENSMUST00000020718.4
|
Smtn
|
smoothelin |
| chr13_-_12520377 | 1.19 |
ENSMUST00000179308.1
|
Edaradd
|
EDAR (ectodysplasin-A receptor)-associated death domain |
| chr1_-_52232296 | 1.19 |
ENSMUST00000114512.1
|
Gls
|
glutaminase |
| chr3_-_92621173 | 1.19 |
ENSMUST00000170676.2
|
Lce6a
|
late cornified envelope 6A |
| chr11_-_99322943 | 1.18 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr2_+_10372426 | 1.17 |
ENSMUST00000114864.2
ENSMUST00000116594.2 ENSMUST00000041105.6 |
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr6_+_134981998 | 1.13 |
ENSMUST00000167323.1
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr10_+_79822617 | 1.13 |
ENSMUST00000046833.4
|
Misp
|
mitotic spindle positioning |
| chr4_+_132351768 | 1.12 |
ENSMUST00000172202.1
|
Gm17300
|
predicted gene, 17300 |
| chr4_-_42853888 | 1.11 |
ENSMUST00000107979.1
|
Gm12429
|
predicted gene 12429 |
| chr1_+_133350510 | 1.11 |
ENSMUST00000094556.2
|
Ren1
|
renin 1 structural |
| chr13_-_98815408 | 1.11 |
ENSMUST00000040340.8
ENSMUST00000099277.4 ENSMUST00000179563.1 ENSMUST00000109403.1 |
Fcho2
|
FCH domain only 2 |
| chr14_+_34086008 | 1.09 |
ENSMUST00000120077.1
|
Anxa8
|
annexin A8 |
| chr5_+_136057138 | 1.09 |
ENSMUST00000111137.1
|
Upk3bl
|
uroplakin 3B-like |
| chr11_-_116168138 | 1.07 |
ENSMUST00000139020.1
ENSMUST00000103031.1 ENSMUST00000124828.1 |
Fbf1
|
Fas (TNFRSF6) binding factor 1 |
| chr14_-_20496780 | 0.96 |
ENSMUST00000022353.3
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr7_+_24884651 | 0.95 |
ENSMUST00000153451.2
ENSMUST00000108429.1 |
Rps19
|
ribosomal protein S19 |
| chr13_-_22035589 | 0.92 |
ENSMUST00000091742.4
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr3_+_59925214 | 0.86 |
ENSMUST00000049476.2
|
C130079G13Rik
|
RIKEN cDNA C130079G13 gene |
| chr16_+_57353093 | 0.85 |
ENSMUST00000159816.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr2_+_86007778 | 0.84 |
ENSMUST00000062166.1
|
Olfr1032
|
olfactory receptor 1032 |
| chr1_+_132880273 | 0.82 |
ENSMUST00000027706.3
|
Lrrn2
|
leucine rich repeat protein 2, neuronal |
| chr16_+_92478743 | 0.81 |
ENSMUST00000160494.1
|
2410124H12Rik
|
RIKEN cDNA 2410124H12 gene |
| chr10_-_130127055 | 0.81 |
ENSMUST00000074161.1
|
Olfr824
|
olfactory receptor 824 |
| chr11_+_118476824 | 0.79 |
ENSMUST00000135383.2
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr4_-_129641060 | 0.79 |
ENSMUST00000046425.9
ENSMUST00000133803.1 |
Txlna
|
taxilin alpha |
| chr19_-_10974664 | 0.77 |
ENSMUST00000072748.6
|
Ms4a10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr15_+_100154379 | 0.74 |
ENSMUST00000023768.6
ENSMUST00000108971.2 |
Dip2b
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr3_-_19265007 | 0.74 |
ENSMUST00000091314.4
|
Pde7a
|
phosphodiesterase 7A |
| chr9_+_5298517 | 0.73 |
ENSMUST00000027015.5
|
Casp1
|
caspase 1 |
| chr7_-_114927726 | 0.73 |
ENSMUST00000059737.2
|
Gm6816
|
predicted gene 6816 |
| chr9_-_100486788 | 0.73 |
ENSMUST00000098458.3
|
Il20rb
|
interleukin 20 receptor beta |
| chr1_+_165485168 | 0.73 |
ENSMUST00000111440.1
ENSMUST00000027852.8 ENSMUST00000111439.1 |
Adcy10
|
adenylate cyclase 10 |
| chr13_+_23574381 | 0.72 |
ENSMUST00000090776.4
|
Hist1h2ad
|
histone cluster 1, H2ad |
| chr1_+_55237177 | 0.69 |
ENSMUST00000061334.8
|
Mars2
|
methionine-tRNA synthetase 2 (mitochondrial) |
| chr7_+_24884611 | 0.68 |
ENSMUST00000108428.1
|
Rps19
|
ribosomal protein S19 |
| chr7_+_75455534 | 0.68 |
ENSMUST00000147005.1
ENSMUST00000166315.1 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr2_+_152962485 | 0.68 |
ENSMUST00000099197.2
ENSMUST00000103155.3 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr5_-_115194283 | 0.66 |
ENSMUST00000112113.1
|
Cabp1
|
calcium binding protein 1 |
| chr10_+_110745433 | 0.65 |
ENSMUST00000174857.1
ENSMUST00000073781.5 ENSMUST00000173471.1 ENSMUST00000173634.1 |
E2f7
|
E2F transcription factor 7 |
| chr14_-_118925314 | 0.64 |
ENSMUST00000004055.8
|
Dzip1
|
DAZ interacting protein 1 |
| chr5_-_6876523 | 0.64 |
ENSMUST00000164784.1
|
Zfp804b
|
zinc finger protein 804B |
| chr11_-_94782500 | 0.62 |
ENSMUST00000162809.2
|
Tmem92
|
transmembrane protein 92 |
| chr6_+_127446819 | 0.62 |
ENSMUST00000112191.1
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr15_-_98004634 | 0.61 |
ENSMUST00000131560.1
ENSMUST00000088355.5 |
Col2a1
|
collagen, type II, alpha 1 |
| chr3_+_64081642 | 0.61 |
ENSMUST00000029406.4
|
Vmn2r1
|
vomeronasal 2, receptor 1 |
| chr6_-_38837224 | 0.59 |
ENSMUST00000160962.1
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr12_-_74316394 | 0.58 |
ENSMUST00000110441.1
|
Gm11042
|
predicted gene 11042 |
| chr14_-_70653081 | 0.55 |
ENSMUST00000062629.4
|
Npm2
|
nucleophosmin/nucleoplasmin 2 |
| chr8_+_10006656 | 0.53 |
ENSMUST00000033892.7
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr10_+_21593151 | 0.52 |
ENSMUST00000057341.4
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr1_-_79858627 | 0.49 |
ENSMUST00000027467.4
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr10_+_56377300 | 0.49 |
ENSMUST00000068581.7
|
Gja1
|
gap junction protein, alpha 1 |
| chr1_+_106861144 | 0.49 |
ENSMUST00000086701.6
ENSMUST00000112730.1 |
Serpinb5
|
serine (or cysteine) peptidase inhibitor, clade B, member 5 |
| chr2_-_127521358 | 0.49 |
ENSMUST00000028850.8
ENSMUST00000103215.4 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr5_+_24100578 | 0.46 |
ENSMUST00000030841.5
ENSMUST00000163409.1 |
Klhl7
|
kelch-like 7 |
| chr3_-_92573715 | 0.45 |
ENSMUST00000053107.4
|
Ivl
|
involucrin |
| chr6_+_128438757 | 0.44 |
ENSMUST00000144745.1
|
Gm10069
|
predicted gene 10069 |
| chr11_+_116593687 | 0.44 |
ENSMUST00000153476.1
|
Aanat
|
arylalkylamine N-acetyltransferase |
| chr4_-_129640959 | 0.43 |
ENSMUST00000132217.1
ENSMUST00000130017.1 ENSMUST00000154105.1 |
Txlna
|
taxilin alpha |
| chr5_+_36204016 | 0.42 |
ENSMUST00000052224.5
|
Psapl1
|
prosaposin-like 1 |
| chr7_+_141079125 | 0.42 |
ENSMUST00000159375.1
|
Pkp3
|
plakophilin 3 |
| chr1_-_74194631 | 0.41 |
ENSMUST00000053389.4
|
Cxcr1
|
chemokine (C-X-C motif) receptor 1 |
| chrX_+_74254782 | 0.41 |
ENSMUST00000119197.1
ENSMUST00000088313.4 |
Emd
|
emerin |
| chr2_+_136891501 | 0.39 |
ENSMUST00000141463.1
|
Slx4ip
|
SLX4 interacting protein |
| chr19_-_47138280 | 0.39 |
ENSMUST00000140512.1
ENSMUST00000035822.1 |
Calhm2
|
calcium homeostasis modulator 2 |
| chr9_-_76567092 | 0.37 |
ENSMUST00000183437.1
|
Fam83b
|
family with sequence similarity 83, member B |
| chr9_-_114982739 | 0.37 |
ENSMUST00000053150.5
|
Gm9846
|
predicted gene 9846 |
| chr7_+_24399921 | 0.36 |
ENSMUST00000108434.1
|
Smg9
|
smg-9 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr15_-_98004695 | 0.35 |
ENSMUST00000023123.8
|
Col2a1
|
collagen, type II, alpha 1 |
| chr8_-_111854278 | 0.34 |
ENSMUST00000034432.5
|
Cfdp1
|
craniofacial development protein 1 |
| chr1_+_51289106 | 0.33 |
ENSMUST00000051572.6
|
Sdpr
|
serum deprivation response |
| chr11_-_95699143 | 0.33 |
ENSMUST00000062249.2
|
Gm9796
|
predicted gene 9796 |
| chr9_+_24097996 | 0.32 |
ENSMUST00000133787.1
ENSMUST00000059650.4 |
Npsr1
|
neuropeptide S receptor 1 |
| chr14_-_49066368 | 0.30 |
ENSMUST00000161504.1
|
Exoc5
|
exocyst complex component 5 |
| chr1_-_193130201 | 0.29 |
ENSMUST00000085555.1
|
Diexf
|
digestive organ expansion factor homolog (zebrafish) |
| chr16_+_20717665 | 0.27 |
ENSMUST00000021405.7
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr10_+_94550852 | 0.27 |
ENSMUST00000148910.1
ENSMUST00000117460.1 |
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr11_-_113209633 | 0.24 |
ENSMUST00000130373.1
|
4732490B19Rik
|
RIKEN cDNA 4732490B19 gene |
| chr4_+_148558422 | 0.23 |
ENSMUST00000017408.7
ENSMUST00000076022.6 |
Exosc10
|
exosome component 10 |
| chr9_+_49518336 | 0.22 |
ENSMUST00000068730.3
|
Gm11149
|
predicted gene 11149 |
| chr10_+_80622677 | 0.22 |
ENSMUST00000079773.6
|
Csnk1g2
|
casein kinase 1, gamma 2 |
| chr1_+_161969284 | 0.22 |
ENSMUST00000160881.1
ENSMUST00000159648.1 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr1_+_161969179 | 0.21 |
ENSMUST00000111594.2
ENSMUST00000028021.6 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr12_-_26415256 | 0.21 |
ENSMUST00000020971.6
ENSMUST00000062149.4 |
Rnf144a
|
ring finger protein 144A |
| chr4_+_112232245 | 0.21 |
ENSMUST00000038455.5
ENSMUST00000170945.1 |
Skint3
|
selection and upkeep of intraepithelial T cells 3 |
| chr16_-_20316750 | 0.20 |
ENSMUST00000182741.1
|
Cyp2ab1
|
cytochrome P450, family 2, subfamily ab, polypeptide 1 |
| chr4_+_42318334 | 0.19 |
ENSMUST00000178192.1
|
Gm21598
|
predicted gene, 21598 |
| chr18_+_66458587 | 0.18 |
ENSMUST00000025399.7
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr19_+_46328179 | 0.18 |
ENSMUST00000026256.2
ENSMUST00000177667.1 |
Fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chr2_+_129100995 | 0.15 |
ENSMUST00000103205.4
ENSMUST00000028874.7 |
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr15_-_98898483 | 0.15 |
ENSMUST00000023737.4
|
Dhh
|
desert hedgehog |
| chr6_-_118562226 | 0.13 |
ENSMUST00000112830.1
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr7_+_24399606 | 0.12 |
ENSMUST00000002280.4
|
Smg9
|
smg-9 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr9_-_106158109 | 0.10 |
ENSMUST00000159809.1
ENSMUST00000162562.1 ENSMUST00000036382.6 ENSMUST00000112543.2 |
Glyctk
|
glycerate kinase |
| chr12_+_107488632 | 0.09 |
ENSMUST00000082269.2
|
3110018I06Rik
|
RIKEN cDNA 3110018I06 gene |
| chr7_-_28962265 | 0.09 |
ENSMUST00000068045.7
|
Actn4
|
actinin alpha 4 |
| chr3_+_79885930 | 0.09 |
ENSMUST00000029567.8
|
Fam198b
|
family with sequence similarity 198, member B |
| chrX_+_136822781 | 0.07 |
ENSMUST00000113085.1
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr7_-_28962223 | 0.07 |
ENSMUST00000127210.1
|
Actn4
|
actinin alpha 4 |
| chr9_-_106656081 | 0.06 |
ENSMUST00000023959.7
|
Grm2
|
glutamate receptor, metabotropic 2 |
| chr5_-_138272786 | 0.06 |
ENSMUST00000161279.1
ENSMUST00000161647.1 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chrX_-_56549606 | 0.03 |
ENSMUST00000081133.4
|
Gm648
|
predicted gene 648 |
| chr2_+_88817173 | 0.03 |
ENSMUST00000072057.1
|
Olfr1202
|
olfactory receptor 1202 |
| chr16_+_91391721 | 0.02 |
ENSMUST00000160764.1
|
Gm21970
|
predicted gene 21970 |
| chr15_-_102246439 | 0.02 |
ENSMUST00000063339.7
|
Rarg
|
retinoic acid receptor, gamma |
| chr7_+_82851963 | 0.01 |
ENSMUST00000179662.1
|
1700010L04Rik
|
RIKEN cDNA 1700010L04 gene |
| chr13_+_41148796 | 0.00 |
ENSMUST00000141292.1
|
Sycp2l
|
synaptonemal complex protein 2-like |
| chr7_+_5034118 | 0.00 |
ENSMUST00000076791.3
|
4632433K11Rik
|
RIKEN cDNA 4632433K11 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp53
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 4.2 | 12.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 3.8 | 11.4 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 2.6 | 10.6 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 1.4 | 7.0 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 1.4 | 5.6 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 1.3 | 5.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 1.0 | 8.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.8 | 6.6 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.8 | 3.3 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.7 | 3.7 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.7 | 2.2 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.7 | 4.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.6 | 5.7 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.5 | 1.9 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.5 | 4.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.4 | 1.7 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.4 | 7.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.4 | 2.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.4 | 1.8 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.3 | 1.6 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.3 | 1.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 | 1.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.3 | 1.4 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.7 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 0.2 | 1.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 0.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 3.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 1.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 2.9 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 3.4 | GO:0099623 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) |
| 0.2 | 3.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 2.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 2.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 2.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 2.2 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 9.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 2.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 1.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.5 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.1 | 0.7 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 2.0 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.1 | 0.3 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 2.0 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.1 | 1.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 3.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 2.8 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 6.1 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 0.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 1.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.4 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 2.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 2.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.4 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 1.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 2.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.0 | 6.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.0 | 4.8 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.9 | 18.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.7 | 5.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.7 | 2.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.7 | 10.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 5.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 6.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.4 | 1.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.3 | 9.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 3.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 7.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 8.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 17.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 7.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 4.4 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 1.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 11.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 4.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0045178 | basal part of cell(GO:0045178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 1.4 | 11.4 | GO:0050786 | Toll-like receptor 4 binding(GO:0035662) RAGE receptor binding(GO:0050786) |
| 1.0 | 8.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 3.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.7 | 2.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.6 | 3.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.6 | 17.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.6 | 2.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.6 | 1.7 | GO:0070737 | protein-glycine ligase activity, elongating(GO:0070737) |
| 0.6 | 2.8 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.6 | 6.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 2.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 4.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 1.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 2.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.3 | 2.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 2.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 12.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 7.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 1.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 2.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 2.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 1.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 6.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 3.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 8.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 2.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 8.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 2.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 7.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 2.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 3.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.7 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 2.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 15.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 4.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 3.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 17.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 6.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 8.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 3.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 9.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 6.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |